- Accueil
- Volume 23 (2019)
- Numéro 4
- Survey of animal by-products in feedingstuffs before the reintroduction of processed animal proteins in aquafeed
Visualisation(s): 0 (0 ULiège)
Téléchargement(s): 0 (0 ULiège)
Survey of animal by-products in feedingstuffs before the reintroduction of processed animal proteins in aquafeed

Notes de la rédaction
The first two authors contributed equally to this work
Received 25 July 2019, accepted 16 September 2019, available online 24 October 2019
This article is distributed under the terms and conditions of the CC-BY License (http://creativecommons.org/licenses/by/4.0)
Résumé
Relevé des sous-produits d’origine animale présents dans l’alimentation des animaux d’élevage avant la réintroduction des protéines animales transformées en aquaculture
Description du sujet. Depuis 2001, le nombre de cas d’encéphalopathie spongiforme bovine a considérablement diminué. Une réintroduction partielle des protéines animales transformées a donc été possible en 2013, mais cette levée partielle rend le contrôle des aliments bien plus complexe.
Objectifs. L’objectif était de donner un aperçu de la situation analytique actuelle et des besoins analytiques actuels et futurs dans le contexte d’une levée progressive de cette interdiction.
Méthode. Des aliments, collectés avant la réintroduction des protéines animales transformées en aquaculture, ont été analysés par microscopie optique puis par PCR afin d’évaluer les difficultés qui pourraient être rencontrées après 2013. Des dosages immunologiques pour la détection de protéines de lait ont également été effectués sur les échantillons contenant de l’ADN de ruminant.
Résultats. En se limitant aux seuls résultats de microscopie, tous les échantillons étaient conformes aux exigences légales. En y ajoutant les résultats de la PCR, l’interprétation devient plus complexe. De l’ADN d’animaux terrestres était en effet détecté dans presque 65 % des cas. Dans les aliments autres que pour les poissons, l’utilisation de produits laitiers, confirmée dans la plupart des cas par les dosages immunologiques, pourrait expliquer la présence d’ADN de ruminant. Dans les aliments pour poissons, 23 % (13/57) des échantillons contenaient de l’ADN ruminant alors que des protéines de lait n’ont pu être détectées que dans seulement trois d’entre eux.
Conclusions. Cette étude a souligné le besoin crucial de solutions analytiques complémentaires afin d’identifier la source de l’ADN détecté.
Abstract
Description. Since 2001, the number of bovine spongiform encephalopathy cases has drastically declined. A gradual lifting of the feed ban for aquaculture was possible in 2013 but this partial relaxation makes its control much more complex.
Objectives. The objective was to have a practical overview of the current analytical situation and to highlight current and foreseen analytical gaps in the context of a progressive lifting of the ban.
Method. Feed samples, collected before the partial lifting as regards aquafeed in 2013, were firstly analyzed by light microscopy. All samples were additionally analyzed by PCR in order to study the difficulties which would be faced with feeds produced after the relaxation. ELISA for the detection of milk proteins was also performed on samples in which ruminant DNA was detected.
Results. When keeping the interpretation to the sole light microscopy results, all samples were in accordance to the legal requirements. Adding the PCR results made the interpretation more difficult. DNA of terrestrial animals was detected in nearly 65% of the samples. Apart from aquafeeds, the presence of ruminant DNA could be explained by the use of dairy products confirmed in most cases by ELISA. In aquafeeds, 23% (13/57) of the samples tested positive for ruminant DNA but only three of them also tested positive for milk proteins.
Conclusions. The study underlined the crucial need for complementary analytical solutions in order to identify the source of the detected DNA.
Table des matières
1. Introduction
1Since the imposition of total ban on processed animal proteins (PAPs) in animal feed, the epidemic situation with regard to bovine spongiform encephalopathy (BSE) has changed and our understanding of the disease has improved. As a result, the legal framework has evolved and animal PAPs have gradually been reintroduced: non-ruminant PAPs and insect PAPs in aquafeed in 2013 (European Commission, 2013) and 2017 (European Commission, 2017) respectively. But with each relaxation, the complexity of the analytical challenge increases in order to meet the two major prohibitions (European Union, 2001; European Commission, 2009a) which remain as pillars: the intra-species recycling and the use of ruminant PAPs in feed. Until 2013, the analytical scheme was in relatively confined situation in which feed analysis by light microscopy (LM) was sufficient. Indeed, whatever the origin of the tissues, if any animal particle was detected in a feed for farmed animals, the feed was considered as containing unauthorised by-products, with some exceptions like the use of fish meal in feed intended for non-ruminants. In July 2013, PAPs of non-ruminant origin were reauthorised in aquafeed, while ruminant PAPs remain banned. This reauthorisation was based on a risk assessment conducted by the European Food Safety Authority (EFSA Panel on Biological Hazards, 2011) and the availability of control tools to ensure the proper implementation of the regulation. For this lifting, the notion of “species of origin” became the main challenge, even if in this case the notion of species was limited to the distinction between ruminant and non-ruminant. Logically, the solution came from the development of a polymerase chain reaction (PCR) method. To date, official controls have been conducted by a combination of LM and PCR methods following operational schemes (European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013a), but future changes will require new analytical methods.
2The present survey was carried out on feeds intended for farmed animal produced before the reauthorisation of non-ruminant PAPs in aquafeed. A total of 86 feed samples were collected from several producers between 2008 and 2012. The goal of this survey was to provide an overview of the situation in compound feed before this lifting. As the aim of this study was to have an extended view of the situation, the two official methods of analysis for the determination of constituents of animal origin in feed (LM and PCR) were performed on all feeds. The samples in which ruminant DNA was detected were also analysed for the detection of milk products with dedicated immunoassays. The interpretation and discussion of the results took into consideration the method of analysis, LM, which was applicable at the time of feed production (European Commission, 2009b) as well as following the operational schemes introduced in 2013 with the reintroduction of non-ruminant PAPs in aquafeed (European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013a). Finally, the complete results were also evaluated for their usefulness for the detection of unauthorised animal by-products.
2. Materials and methods
2.1. Samples
3Eighty-six compound feeds were collected from European feed industries and agencies between 2008 and 2012. The feeds were classified into three groups: 20 feeds were described as intended for cattle, ruminant or non-ruminant animals, 57 feeds were intended for fish feeding and the destination of 9 feeds was not specified. All samples were kept at 4 °C, and subsamples were ground at 2 mm with a rotor mill (ZM200, Retsch, Haan, Germany). For PCR analysis, subsamples were submitted to a second grinding with a 0.5 mm sieve when the samples contained a level of fat allowing such fine grinding. Between each grinding, the grinder was disassembled and all the pieces were cleaned with detergent, decontaminated in bleach bath for 15 min and rinsed with water.
2.2. Light microscopy (LM)
4All samples were analysed in duplicate. Analyses were performed according to Annex VI of Commission Regulation (EC) No 152/2009 amended by Commission Regulation (EC) No 51/2013 (European Commission, 2013). After at least 5 min settling time, the sediment was recovered in a filter paper by opening the stopcock and air-dried. After removing the tetrachloroethylene from the funnel by opening the stopcock, the funnel was inverting to recover the flotate onto a Petri dish. It was dried in a laboratory fume hood at room temperature. The sediment was stained using Alizarin Red S and then sieved with a mesh size of 250 µm in order to obtain a coarse fraction > 250 µm and a fine fraction < 250 µm.
5The coarse and fine fractions of the sediment were then mounted on slides using Norland Optical Adhesive 65® (Norland Products Inc., Cranbury, USA) embedding agent in order to make them permanent by applying ultraviolet light curing (Veys & Baeten, 2010; European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013b). Flotate was mounted using appropriate mounting reagents (e.g. Fehling reagents, cystine reagents, aqueous lugol). All slides were observed using transmitted light microscopy under brightfield conditions.
2.3. Polymerase chain reaction (PCR)
6Genomic DNA was extracted from the samples following the method recommended in the EURL-AP Standard Operating Procedure (European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013c) using the “Wizard® Magnetic DNA Purification System for Food” kit (Promega, Madison, WI, USA). Test portions of 100 mg were weighed and all samples were analysed in duplicate. Primers and probes used in this study for ruminant (European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013d) and chicken (Fumière et al., 2010) DNA detection are listed in table 1. The porcine method used was successfully validated through an inter-laboratory study conducted by the EURL-AP in 2015 but the results are not yet publicly available. The primers and probes for fish DNA detection are unpublished. The oligonucleotides were synthesised by Eurogentec (Seraing, Belgium). The probes were labelled with the reporter dye FAM™ (6-carboxyfluorescein) at the 5’end and the quencher dye TAMRA™ (Tetramethyl-6-Carboxyrhodamine) at the 3’end.
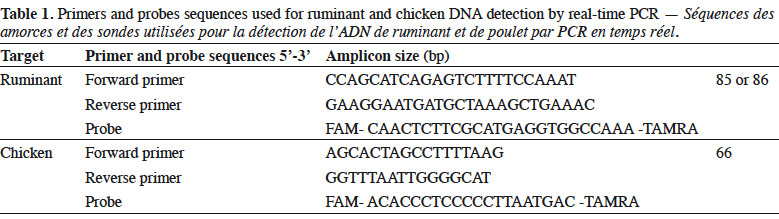
7Real-time PCR was performed on a thermocycler LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using the Universal Master Mix of Diagenode (Liège, Belgium). The reaction mixtures used are presented in table 2.
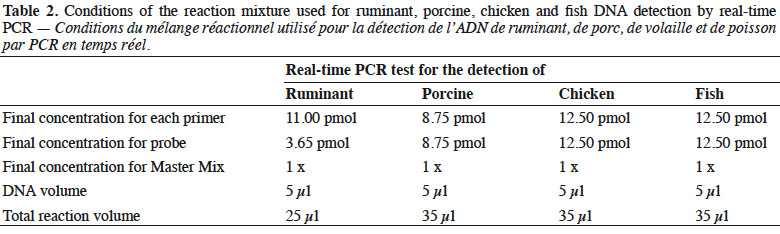
8The thermal programmes were the following: the first stage was performed at 50 °C for 2 min for the activation of uracil-N-glycosylase (UNG), the second stage consisted of 95 °C for 10 min in order to inactivate the UNG, activate the hot start polymerase and denature the DNA template, while the last stage consisted of 50 amplification cycles, each including a denaturation step at 95 °C for 15 s followed by an annealing/elongation step for 60 s at 60 °C for the ruminant PCR test and 50 °C for the porcine, chicken and fish PCR test.
2.4. Immunoassays
9Analyses were outsourced to the CER Groupe (http://www.cergroupe.be/fr/) by using enzyme-linked immunosorbent assays (ELISA). These assays were initially developed for the detection of milk allergens in food matrices with a limit of quantification (LOQ) for beta-lactoglobulin (BLG) and casein of 0.25 ppm and 0.5 ppm, respectively (Dumont et al., 2010).
3. Results
3.1. Compound feed intended for farmed animals other than aquaculture and fur animals
10By LM, all samples gave negative results for the detection of animal proteins from terrestrial origin and 9 out of 20 were found to contain animal particles from fish origin (Table 3).
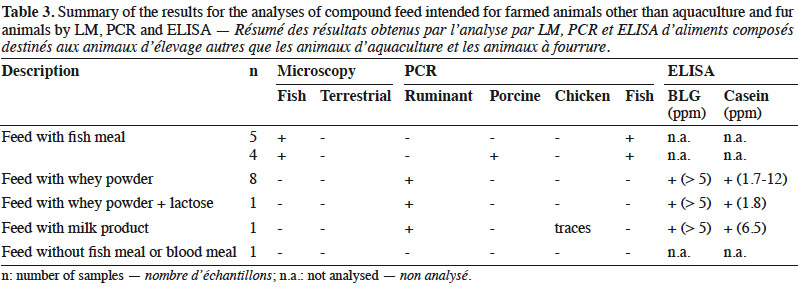
11Regarding PCR results, 10 out the 20 samples gave positive signals for ruminant DNA, whereas 4 samples contained porcine DNA and, as expected, fish DNA was detected in 9 samples. In one sample, traces of chicken DNA were also detected.
12The 10 samples containing ruminant DNA were also analysed for the presence of milk proteins and gave positive ELISA results for both BLG and casein.
3.2. Compound feed with undetermined destination
13By LM, all samples gave negative results for the detection of animal proteins of terrestrial and fish origin respectively (Table 4).
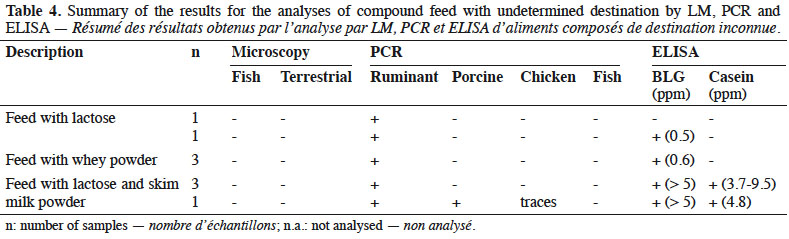
14With the help of PCR, ruminant DNA was detected in all feeds whereas search for other specific animal targets (porcine, chicken and fish DNA) gave a negative result, except for one sample which gave a positive result for porcine DNA and in which traces of chicken DNA were also observed.
15All samples were analysed by ELISA: eight samples were positive for BLG and four of these were also positive for casein.
3.3. Compound feed intended for aquaculture animals
16By LM, all samples gave negative results for the detection of animal proteins of terrestrial origin and all were found to contain animal proteins of fish origin (Table 5).
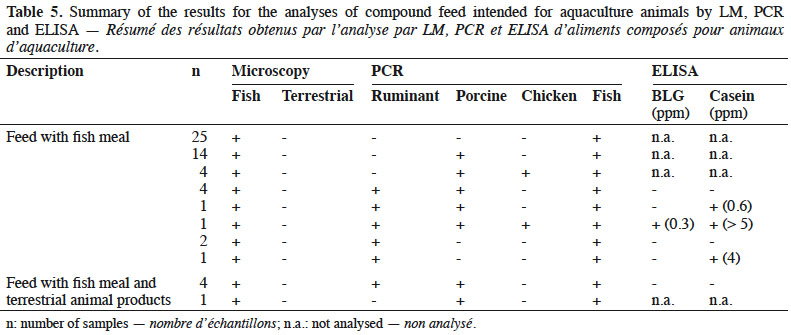
17The PCR results showed all samples to be positive for fish DNA. The results obtained with the other targets (ruminant, porcine and chicken DNA) were analysed separately depending on the labelling:
18– fish feed without any mention of the use of terrestrial products: 48% (25/52) of these feeds were negative for all other targets (ruminant, porcine and chicken DNA). In 46% (24/52) of these samples, porcine DNA was identified and chicken DNA was also detected in 10% (5/52) of them. Ruminant DNA was identified in 17% (9/52) of the samples. The identification of ruminant DNA was combined in 33% of the cases with the detection of at least one milk protein by ELISA.
19– fish feed containing terrestrial animal products: for five fish feeds, the use of terrestrial animal products was specified in the labelling. For four of these, the use of blood meal was specified and for one, no more details were given. All of them were positive for the presence of porcine DNA. Eighty percent of these feeds (four out of five) were also positive for the presence of ruminant DNA, but none of them tested positive for the presence of milk proteins with ELISA. No chicken DNA was detected.
4. Discussion
4.1. Compound feed intended for farmed animals other than aquaculture and fur animals
20LM results were in line with the labelling as the nine positive samples for fish were known to contain fish meal as feed material (Table 3). Therefore, from a legal point of view, all feeds complied with the requirements.
21Following the operational schemes introduced in 2013, LM was sufficient to detect the presence of prohibited constituents of animal origin in these types of feed. However, PCR analyses were conducted in order to evaluate the presence of any undesired or unexplained signal. The signals for the presence of ruminant DNA were easily explained by the labelling as these feeds were known to contain dairy products (whey powder, lactose or milk products). This assumption was confirmed by a positive ELISA result for both BLG and casein. However, although the presence of ruminant DNA seems to be linked to the use of dairy products, an additional presence of prohibited animal by-products of ruminant origin such as bovine blood meal could not be excluded (Lecrenier et al., 2018). The positive results for fish DNA matched to the samples in which fish particles were identified. The origin of the porcine DNA is more difficult to determine. In 25% of the samples in which porcine DNA was detected, animal fat was used as feed material. For the other samples, no indication explained these results. As no particles of terrestrial animals were detected by LM, one hypothesis is that the presence of porcine DNA was due to porcine by-products without any typical and morphologically identifiable characteristics identified by LM such as blood meal, blood products or gelatine.
22These results showed that PCR or ELISA could provide additional information to the LM results and help with the interpretation of the results. Feed materials of porcine origin seem to be used in non-ruminant feeds (feed for chicken in these cases) but the type of by-products could not be determined with the implemented analytical methods.
4.2. Compound feed with undetermined destination
23Although the destination of the feed was not declared, the absence of animal proteins microscopically detectable (Table 4) made these feeds suitable to feed farmed animals.
24All these feeds were known to contain dairy products (whey powder, milk powder or lactose). This may explain the results obtained by PCR for ruminant DNA detection. ELISA results confirmed this point. Here again, the additional presence of prohibited animal by-products of ruminant origin such as bovine blood meal cannot be excluded. For one sample, the origin of the detected ruminant DNA cannot be explained. It is known that lactose is used in this feed and it can be assumed that this sugar, which is a by-product of the dairy industry, is the origin of the ruminant DNA without any presence of milk proteins. Finally, the origin of porcine DNA in one sample could not be clarified.
25In summary, for two samples, the use of prohibited animal by-products can be considered. Unfortunately the current analytical methods are insufficient to reject or confirm this suspicion.
4.3. Compound feed intended for aquaculture animals
26LM and PCR results for fish DNA (Table 5) were in line with the labelling as all labels declared the use of fish meal as feed material. From a legal point of view, all feeds complied with the legal requirements before the reintroduction of non-ruminant PAPs in aquafeed.
27The results obtained with the other PCR targets (ruminant, porcine and chicken DNA) are clearly difficult to interpret because of a crucial lack of information. Regarding the fish feeds without any mention of the use of terrestrial products, two main conclusions can be drawn:
28– a high percentage (46%) of fish feeds were positive for porcine DNA;
29– the presence of ruminant DNA could not be explained in 66% of the samples by the presence of milk despite the high sensitivity of the ELISA method.
30For the five fish feed containing terrestrial animal products, although the positive results for porcine DNA can be easily explained by the labelling (use of terrestrial animal products), the positive results for ruminant DNA remained totally unexplained.
31In total, 56% (32/57) of the fish feeds contained DNA of terrestrial animals and most of them tested positive at least for porcine DNA (29/57). This reveals the wide use of porcine by-products in aquafeed. As no bones of terrestrial animals were detected, the DNA probably originated from porcine blood meal, blood products or gelatine. All these products were authorised at the time of production.
32The origin of the ruminant DNA in 23% (13/57) of the fish feeds was more difficult to interpret. Three of these fish feeds also tested positive for milk proteins with ELISA. Although the use of milk may explain for the presence of ruminant DNA, it cannot be excluded that unauthorised ruminant by-products were also present. Moreover, it can be assumed that ruminant DNA in ten out of these thirteen samples that tested negative for milk protein originated from unauthorised by-products. Besides, in 77% (10/13) of the cases, the presence of ruminant DNA was associated with the presence of porcine DNA, suggesting a possible contamination of porcine blood meal or blood products with ruminant products.
33Only 5/57 of the samples tested positive for chicken DNA. Although its origin is unknown, the use of chicken by-products seems to be limited.
5. Conclusions
34Since 1986, more than 190,000 cases of bovine spongiform encephalopathy (BSE) have been reported in Europe (EFSA, 2017). Even today, many questions remain about this disease, offering no more than hypotheses for answers. Uncertainties still exist in relation to the pathogenesis and epidemiology of BSE (Simmons et al., 2017). Occasional cases of classical BSE on animals born after the reinforcement of the feed ban (BARB cases) still occur (EFSA Panel on Biological Hazards [BIOHAZ], 2017). It is not clear whether these cases are due to an incorrect implementation of the feed ban or to spontaneous incidents as the small number of cases does not support a robust risk assessment.
35These cases remind us that if this disease is not fully understood it is important to remain cautious, otherwise a new breaking out is always possible. The difficulty of diagnosis currently still impossible in ante-mortal (alive animal) and the zoonotic nature of BSE, make the prevention role of the specified risk materials removal and the feed ban of paramount importance.
36In this survey, various types of feed were analysed using the official methods. In a first approach, by limiting the interpretation of the results to the LM analyses as provided by the legislation in force at that time (i.e. before June 2013), all samples were in accordance to the legal requirements with no particles identified from terrestrial animals. Nevertheless, by adding the PCR results, it was demonstrated that, when analyses were limited to LM, some information that might lead to different interpretations was missing. The presence of DNA of terrestrial animals was detected in nearly 65% of the samples. The first conclusion that can be drawn is that products of porcine and ruminant origin were often present in animal feed while the presence of chicken DNA was quite rare and always associated with the presence of DNA of other terrestrial animals.
37Apart from aquafeeds, the presence of ruminant DNA could be explained by the use of dairy products confirmed in most cases (18/19) by ELISA tests. In aquafeeds, 23% (13/57) of the samples tested positive for ruminant DNA but only three of these fish feeds also tested positive for milk proteins with ELISA. Although ELISA helped with the interpretation of some cases, it must be kept in mind that the presence of authorised product does not exclude the presence of unauthorised ones (e.g. ruminant blood meal, ruminant blood products). It is especially important to take this point into consideration as fraudsters could take advantage of analytical limitations by using authorised by-products to hide banned by-products.
38Regarding the detection of porcine DNA, in most cases its origin could not be determined. As no particles of terrestrial animals were detected, one hypothesis was the presence of porcine DNA due to porcine by-products undetectable by LM (e.g. blood meal, blood products) or without any microscopic characteristics (e.g. such as gelatine). Since the regulation is different for each of these products, this situation clearly underlines the need for a direct method capable of jointly determining both the species and the tissue of an animal by-product included in feed.
39This survey was conducted on samples collected before the reintroduction of non-ruminant PAPs in fish feed. If the same survey had been conducted on samples collected now, interpretation would be even more complex because presence of bones of terrestrial animals is no longer sufficient to consider the sample as non-compliant. If bones of terrestrial animals and DNA from both ruminants and pigs were detected, it would be impossible to determine if the bones were the “marker” of the presence of ruminant PAPs or porcine PAPs and so no firm conclusions could be drawn.
40This study underlined the analytical gaps and the crucial need for analytical methods allowing a combined and simultaneous determination of species and tissue of origin. Indeed, currently, there is no official method that can answer this question as the presence of milk proteins cannot exclude the simultaneous presence of unauthorised products. With the prospect of future reintroduction of non-ruminant PAPs in non-ruminant feed, these rare cases will become frequent. Indeed, whey powder is known to be used in piglet feed for its positive effect on feed intake and gut health (Boudry et al., 2008). Moreover, a prospective study on poultry feed (data not shown) has shown that low level of milk proteins can be found in poultry feed, probably due to the use of lactose as prebiotics to promote the lactobacilli activity (Cesari et al., 2014) or as carrier of some feed additives. Based on this assessment of the analytical situation, novel analytical methods are currently investigated. One of the most promising one is mass spectrometry (MS). Indeed, MS is a method of choice to fill in this analytical gap as it provides simultaneous information about the tissue and species of origin (Lecrenier et al., 2016; Lecrenier et al., 2018). Next to the development of novel methods, some legal concepts (e.g. what is meant precisely by poultry) should be defined more precisely as discussed by van Raamsdonk et al. (2019) to better bridge legal requirements and performance of analytical methods.
Acknowledgements
41The authors wish to thank the members of the EURL-AP team for their technical assistance, with special mention of S. Brichard, M. Collard, J. Hulin, J. Maljean and B. Scaut. They also thank the members of the Food and Feed Quality Unit and the Authentication and Traceability Unit at the Walloon Agricultural Research Centre for their helpful collaboration.
Bibliographie
Boudry C. et al., 2008. Effect of a bovine colostrum whey supplementation on growth performance, faecal Escherichia coli population and systemic immune response of piglets at weaning. Animal, 2, 730-737.
Cesari V. et al., 2014. Egg quality and productive performance of laying hens fed different levels of skimmed milk powder added to a diet containing Lactobacillus acidophilus. Poultr. Sci., 93, 1197-1201.
Dumont V. et al., 2010. Development of milk and egg incurred reference materials for the validation of food allergen detection methods. Qual. Assur. Saf. Crops Foods, 2, 208-215.
EFSA, 2017. Scientific report on the European Union summary report on surveillance for the presence of transmissible spongiform encephalopathies (TSE) in 2016. EFSA J., 15, 1-68.
EFSA Panel on Biological Hazards, 2011. Scientific opinion on the revision of the quantitative risk assessment (QRA) of the BSE risk posed by processed animal proteins (PAPs). EFSA J., 9, 1947.
EFSA Panel on Biological Hazards (BIOHAZ), 2017. Bovine spongiform encephalopathy (BSE) cases born after the total feed ban. EFSA J., 15, 1-45.
European Commission, 2009a. Regulation (EC) No 1069/2009 of 21 October 2009 laying down health rules as regards animal by-products and derived products not intended for human consumption and repealing Regulation (EC) No 1774/2002 (Animal by-products Regulation). Off. J. Eur. Union, L300, 1-33.
European Commission, 2009b. Commission Regulation (EC) No 152/2009 of 27 January 2009 laying down the methods of sampling and analysis for the official control of feed. Off. J. Eur. Union, L54, 1-130.
European Commission, 2013. Commission Regulation (EU) No 51/2013 of 16 January 2013 amending Regulation (EC) No 152/2009 as regards the methods of analysis for the determination of constituents of animal origin for the official control of feed text with EEA relevance. Off. J. Eur. Union, L20, 33-43.
European Commission, 2017. Commission Regulation (EU) 2017/893 of 24 May 2017 amending Annexes I and IV to Regulation (EC) No 999/2001 of the European Parliament and of the Council and Annexes X, XIV and XV to Commission Regulation (EU) No 142/2011 as regards the provisions on processed animal protein. Off. J. Eur. Union, L138, 92-116.
European Union, 2001. Regulation (EC) No 999/2001 of the European Parliament and of the Council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Off. J. Eur. Union, L147, 1-40.
European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013a. EURL-AP Standard operating procedure - Operational protocols for the combination of light microscopy and PCR, http://eurl.craw.eu/img/page/sops/EURL-AP%20SOP%20operational%20schemes%20V3.0.pdf, (09/08/2018).
European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013b. EURL-AP Standard operating procedure - Slide preparation and mounting, http://eurl.craw.eu/img/page/sops/EURL-AP%20SOP%20slide%20mounting%20V1.0.pdf, (08/08/2018).
European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013c. EURL-AP Standard operating procedure - DNA extraction using the “Wizard® Magnetic DNA purification system for Food” kit, http://eurl.craw.eu/img/page/sops/EURL-AP%20SOP%20DNA%20extraction%20V1.1.pdf, (09/08/2018).
European Union Reference Laboratory for Animal Proteins in feedingstuffs, 2013d. EURL-AP Standard operating procedure - Detection of ruminant DNA in feed using real-time PCR, http://eurl.craw.eu/img/page/sops/EURL-AP%20SOP%20Ruminant%20PCR%20V1.0.pdf, (19/10/2015).
Fumière O. et al., 2010. Development of a real-time PCR protocol for the species origin confirmation of isolated animal particles detected by NIRM. Food. Addit. Contam. Part A, 27, 1118-1127.
Lecrenier M.C. et al., 2016. Identification of specific bovine blood biomarkers with a non-targeted approach using HPLC ESI tandem mass spectrometry. Food Chem., 213, 417-424.
Lecrenier M.C. et al., 2018. A mass spectrometry method for sensitive, specific and simultaneous detection of bovine blood meal, blood products and milk products in compound feed. Food Chem., 245, 981-988.
Simmons M. et al., 2017. Discontools: identifying gaps in controlling bovine spongiform encephalopathy. Transboundary Emerging Dis., 65, 9-21.
van Raamsdonk L. et al., 2019. Bridging legal requirements and analytical methods: a review of monitoring opportunities of animal proteins in feed. Food. Addit. Contam. Part A, 36, 46-73.
Veys P. & Baeten V., 2010. New approach for the quantification of processed animal proteins in feed using light microscopy. Food. Addit. Contam. Part A, 27, 926-934.






