- Startpagina tijdschrift
- Volume 22 (2018)
- Numéro 1
- Cloning of a novel gene from Penicillium oxalicum I1 which in Escherichia coli enhances the secretion of acetic acid
Weergave(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Cloning of a novel gene from Penicillium oxalicum I1 which in Escherichia coli enhances the secretion of acetic acid

Nota's van de redactie
Received 5 May 2017, accepted 4 December 2017, available online 6 February 2018.
This article is distributed under the terms and conditions of the CC-BY License (http://creativecommons.org/licenses/by/4.0)
Résumé
Clonage d’un nouveau gène de Penicillium oxalicum I1 entrainant la sécrétion d’acide acétique chez Escherichia coli
Description du sujet. Les acides organiques jouent un rôle essentiel dans la solubilisation de certains ions dans le sol. La surexpression hétérologue d’un gène unique au sein d’une cellule est une stratégie optimale pour accroitre la sécrétion d’acides organiques solubilisant le phosphate.
Objectifs. Lors de cette étude, nous avons élaboré une banque d’ADNc de Penicillium oxalicum I1. La capacité à solubiliser le phosphate d’un milieu au phosphate tricalcique (TCP) a été éprouvée pour les divers clones ainsi générés car via leur expression dans Escherichia coli, nous avons pu vérifier quels étaient ceux qui engendraient une augmentation de la sécrétion d’acides organiques.
Méthode. Une banque primaire d’ADNc de Penicillium oxalicum I1 a été construite par le mécanisme du « template switching polymerase chain reaction ». La capacité d’E. coli DH5α™ à sécréter des acides organiques par surexpression du gene du clone I-2 de P. oxalicum I1 a été éprouvée dans le milieu TCP comprenant le glucose comme seule source de carbone. Ensuite, on a substitué au glucose les acides pyruvique, citrique α-cétoglutarique, succinique, fumarique ou malique comme seule source de carbone du milieu TCP afin de vérifier à nouveau la capacité de sécrétion d’acides organiques des transformants d'E. coli DH5α™.
Resultats. Un total de 106 clones testés a engendré un halo sur milieu TCP gélosé. Parmi ceux-ci, seul le clone I-2 a fourni un halo clair. L’ADNc du clone I-2 s’étend sur une longueur de 1 151 pb et renferme une phase de lecture ouverte de 702 pb codant pour une protéine hypothétique de 233 acides aminés. La séquence de l’ADNc présente une identité de 68 % ainsi qu’un « query coverage » de 73 % avec des séquences de gènes fongiques dont la fonction demeure inconnue. Le transformant d’E. coli DH5α™ porteur du clone I-2 sécrète jusqu’à 567 mg·l-1 d’acide acétique en 48 h. L’emploi de glucose ou des acides pyruvique, α-cétoglutarique et malique comme seule source de carbone accroit la sécrétion d’acide acétique du transformant. À l’inverse, le recours aux acides citrique, succinique ou fumarique comme seule source de carbone n’améliore pas la sécrétion du clone I-2 comparé à un témoin porteur du seul vecteur pBluescript.
Conclusions. Nous avons mis en évidence un nouveau gène de P. oxalicum I1 dont la surexpression dans E. coli DH5α™ peut entrainer un accroissement de la sécrétion d’acide acétique. Cette observation doit aider à cerner le rôle du gène isolé de P. oxalicum et de ses homologues identifiés dans d’autres espèces du genre Penicillium.
Abstract
Description of the subject. Organic acids play an important role in the conversion of insoluble ions into soluble ones in soil. Heterologous overexpression of a single gene in a cell is the optimal strategy for increasing the secretion of organic acids solubilizing phosphate.
Objectives. In this study, we constructed a primary cDNA library of Penicillium oxalicum I1, and screened clones that can solubilize P in tricalcium phosphate (TCP) medium. We aimed to obtain the gene expressed in Escherichia coli, which can enhance organic acid secretion.
Method. A primary cDNA library of Penicillium oxalicum I1 was constructed using the switching mechanism at the 5'-end of RNA transcription. The organic acid secretion ability of E. coli DH5α™ with overexpressed P. oxalicum I1gene was tested in TCP medium where glucose is the sole carbon source. Afterwards, pyruvic acid, citric acid, α-ketoglutaric acid, succinic acid, fumaric acid, and malic acid were used as sole carbon source substitutes for glucose in the TCP medium to test the organic acid secretion ability of the transformed E. coli DH5α™.
Results. A total of 106 clones showed halos in TCP medium, among which clone I-2 displayed clear halo. The full-length cDNA of clone I-2 was 1,151 bp, with a complete open reading frame of 702 bp, which encoded a hypothetical protein of 233 amino acids. The cDNA sequence showed 68% identity and 73% query cover with other fungal gene sequences of which the function remains unknown. Escherichia coli containing the cloned gene secreted up to 567 mg·l-1 acetic acid within 48 h. The use of glucose, pyruvic acid, α-ketoglutaric acid, and malic acid improved the acetic acid secretion of the E. coli DH5α™ clone I-2. By contrast, the use of citric acid, succinic acid, and fumaric acid did not improve the acetic acid secretion of clone I-2 compared to a control E. coli DH5α™ strain bearing only the cloning vector without any insert.
Conclusions. We obtained a novel gene from Penicillium oxalicum I1 whose overexpression in E. coli DH5α™ increased the secretion of acetic acid. This observation should help to understand what is the function of the gene isolated from P. oxalicum as well as that of its homologs found in several other species of the Penicillium genus.
Inhoudstafel
1. Introduction
1Many soil microorganisms can secrete organic acids, such as oxalic, lactic, acetic, propionic, malic, tartaric, citric, butyric, malonic, succinic, gluconic, and fumaric acids (Banik & Dey, 1982; Altomare et al., 1999; Fomina et al., 2005; Khan et al., 2007; Bianco & Defez, 2010; Gulati et al., 2010). These acids play important roles in agriculture by improving for instance phosphate solubilization and releasing metal ions to improve the concentration of plant essential nutrients (Dessureault-Rompré et al., 2007; Singh et al., 2007). Organic acids can chelate Fe3+, Fe2+, Ca2+, and Al3+, thereby converting insoluble forms of nutrients to soluble ones (Walpola & Yoon, 2013). These acids could decrease fertilizer requirement and improve fertilizer utilization. However, they are scarcely found and the effects are limited in soil. Therefore, highly efficient expression of genes related to exogenous organic acid secretion for organic acid production is an optimal strategy for improving fertilizer utilization. In this strategy, the critical step is obtaining genes that can improve organic acid secretion and expressing these genes heterologously. Numerous studies on genes that can enhance organic acid secretion have been conducted. Genes in bacteria are primarily cloned to improve gluconic acid secretion (Goldstein et al., 1999). For example, pyrroloquinoline quinone genes from Enterobacter intermedium, Klebsiella pneumoniae, and Rahnella aquatilis have been expressed in Escherichia coli (Meulenberg et al., 1992; Kim et al., 1998; Kim et al., 2003). Few fungal genes demonstrate this function as well. Fungal genes have been expressed in E. coli to improve various organic acid secretions. Lü et al. (2012) cloned mitochondrial malate dehydrogenase from Penicillium oxalicum and expressed the gene in E. coli to improve malate, lactate, acetate, citrate, and oxalate secretions. Gong et al. (2014a) also cloned delta-1-pyrroline-5-carboxylate dehydrogenase from P. oxalicum and expressed the gene in E. coli to enhance the secretion of acetic acid and α-ketoglutarate.
2Penicillium oxalicum I1 is a fungus that secretes oxalic acid. In the current study, we constructed a primary cDNA library of P. oxalicum and screened clones that can solubilize phosphate in tricalcium phosphate (TCP) medium by clear halos. We aimed to obtain the gene expressed in E. coli that can enhance organic acid secretion.
2. Materials and methods
2.1. Strains, plasmids, and media
3Penicillium oxalicum I1 was grown at 30 °C in potato dextrose agar medium. Plasmid transformants of E. coli DH5α™ were grown at 37 °C in Luria–Bertani (LB) medium and TCP containing 100 μg·ml-1 ampicillin. The TCP medium comprised the following (per liter of distilled water): glucose, 10 g; 0.3 g of MgSO4·7H2O; 0.3 g of NaCl;0.3 g of KCl; 0.5 g of (NH4)2SO4; 0.03 g of FeSO4·7H2O; 0.03 g of MnSO4·4H2O; and 5 g of Ca3(PO4)2, 15 g of agar (liquid medium without agar) (Nautiyal, 1999). The vector used was pBluescript II SK(+) (TaKaRa, Japan).
2.2. Construction of cDNA library and screening for genes enhancing secretion of organic acid
4A cDNA library of P. oxalicum I1was constructed using the SMART method. The cDNA sequences were linked to pBluescript II SK(+) and recombinant plasmids were transformed into competent E. coli DH5α™ cells. Colonies diluted to 100-fold were spread on TCP medium agar plates containing 100 μg·ml-1 ampicillin. Clear halos appeared after three days. These halos resulted from the solubilization of TCP by the clones secreting organic acid, and the clones were subcultured to confirm stability. The cDNA sequences were blasted in the GenBank database.
5The open reading frame (ORF) was estimated based on the cDNA sequences translated into protein by DNAMAN V6.0. The ORF sequence was amplified with the sense (5'-TATTCGGAATTCATGCAG CCCTCCTACAATGT-3') and antisense (5'-CACGGACTCGAGCTATCGT GTCAAATCTTTCCACAGA-3') primers. The PCR mix contained 1 µM of each primer, 10 µM deoxynucleotide, 10× buffer (NEB, USA), and 1 unit of Taq Polymerase (NEB). This process was conducted under the following thermocycling conditions: initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 60 s, 60 °C for 60 s, 72 °C for 60 s, and a final elongation at 72 °C for 10 min (ABI 9700). The ORF sequence and pBluescript II SK(+) vector were digested with EcoRI and XhoI enzymes at 37 °C for 4 h. Digested products were linked to pBluescript II SK(+) by T4 DNA ligase. Plasmids containing the ORF sequence were transformed into E. coli DH5α™ competent cells. Transformants showing clear halos were selected from the TCP plates.
2.3.Organic acid secretion
6The organic acid secretion ability of E. coli DH5α™ with overexpressed P. oxalicum I1gene was tested in liquid TCP medium. Escherichia coli DH5α™ containing pBluescript II SK(+) and ORF sequence (pPos) or pBluescript II SK(+) (pBlu)were grown in TCP. An aliquot (100 µl) of each bacterial culture (108 cfu·ml-1) was grown in 50 ml of broth. The culture was shaken at 120 rpm at 37 °C. Changes in pH of the medium and organic acid concentrations were measured in the culture filtrates at 0, 8, 16, 24, 36, and 48 h. The pH of the medium was measured using a pH meter equipped with a glass electrode. The cell density of E. coli DH5α™ was tested by plate counting at 24 h and 48 h. Organic acids produced were determined at 48 h by an anion chromatographic system (ICS-3000, Dionex, USA).The experiment was performed three times.
2.4. Substrate utilization
7Pyruvic acid, citric acid, α-ketoglutaric acid, succinic acid, fumaric acid, and malic acid were used as sole carbon source substitutes for glucose in the TCP medium whose pH was adjusted to 7.0. An aliquot (100 µl) of E. coli DH5α™ containing pPos culture (108 cfu·ml-1) was grown in 50 ml of broth. The culture was shaken at 120 rev·min-1 at 37 °C. Organic acids produced were determined at 48 h by an anion chromatography system (ICS-3000, Dionex, USA). The experiment was performed three times.
2.5. Statistical analysis
8Statistical analysis was conducted by using Analysis of Variance (ANOVA) statistical package for social sciences (SPSS) software, version 21.0 followed by comparison of multiple treatment levels with the control, using the significant difference (LSD) at p ≤ 0.05.
3. Results
3.1. Screening for genes enhancing of secretion of organic acids
9Selection of the E. coli transformants with cDNA from P. oxalicum were tested on their ability to produce larger amounts of organic acid by checking the clear halos they induce on TCP agar plates because of the phosphate solubilization by the clones after 3 d of incubation at 37 °C. The titer of the primary cDNA library was 5.65 × 106 cfu·ml-1 with a recombination rate of 99.15%. A total of 106 positive clones were obtained, and the diameters of the clear halos ranged from 1.3 mm to 4.2 mm. The diameter of the clear halo of the clone I-2 was 3.6 mm. The full-length cDNA of the clone I-2 was 1,151 bp, and this clone contained an ORF of 702 bp (pPos). This ORF encoded a 25.41 kDa polypeptide comprising 233 amino acids. The cDNA sequence was deposited to the GenBank under accession number JF419552.
10The cDNA sequence was analyzed through the GenBank database. Pennicillium oxalicum I1 cDNA showed 68% identity and 73% query cover with Penicillium chrysogenum Wisconsin 54-1255 partial mRNA (accession number XM_002569279.1). The sequence from P. oxalicum I1 also showed < 66% identity and 53% query cover with other fungi. The deduced amino acid sequence of P. oxalicum I1 was analyzed using the Blast program (DNAMAN 6.0) via the GenBank database. Multiple alignment results revealed 100% identity between the amino acid sequences of P. oxalicum I1 and P. oxalicum 114-2 (EPS25113.1), which has an unknown gene function. In addition, the sequence from P. oxalicum I1 can be aligned with hypothetical proteins of other closely related fungi (Figure 1). Even though for some the identity is below 63% identity, the alignment clearly shows that these hypothetical proteins belong to a same group that should share the same function.
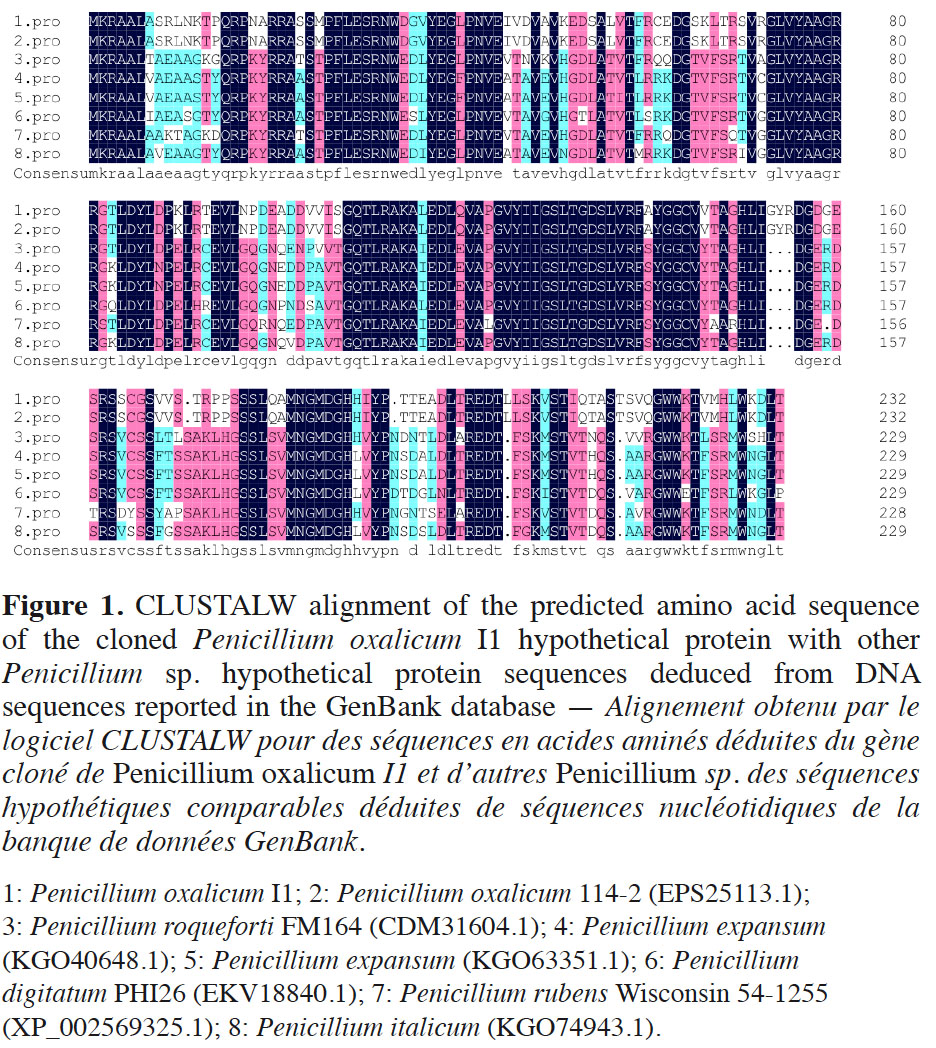
3.2. Organic acid secretion
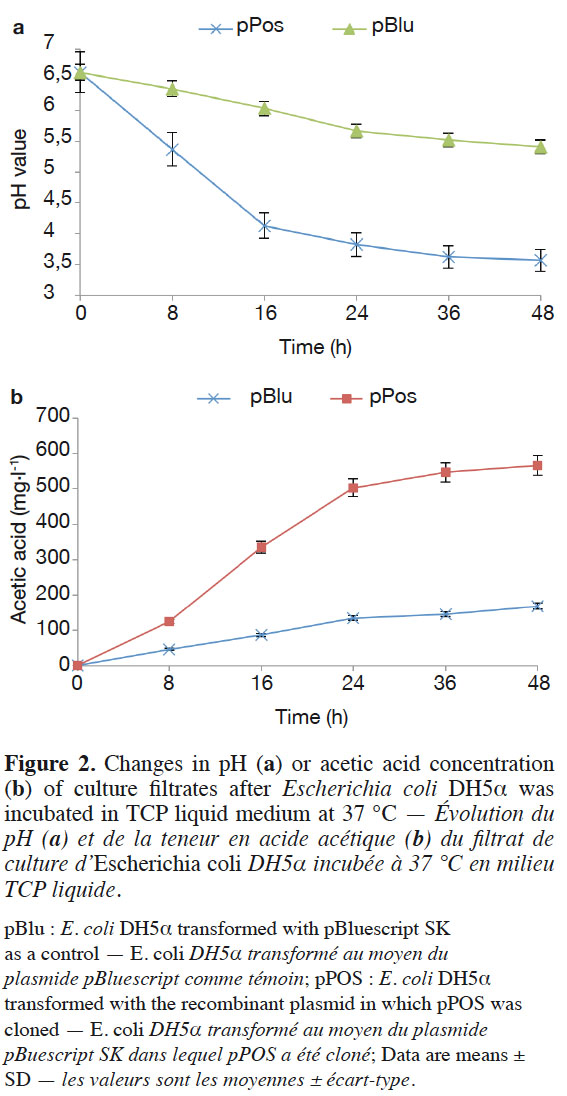
11The subcloned ORF of the cDNA clone was inserted into pBluescript II SK(+), and the resulting recombinant plasmid pPos was transformed into E. coli DH5α™. Clear halos resulting from TCP solubilization appeared in vitro. Finally, E. coli DH5α™ containing the target gene was grown in TCP liquid medium to demonstrate the organic acid secretion ability of the recombinant. The pH values of the TCP liquid medium were more quickly reduced in E. coli bearing the cloned pPos (Figure 2a). The pH level decreased from pH 7.0 to pH 3.57 by pPos after 48 h. Under the same condition, pH level decreased from pH 7.0 to pH 5.41 by pBlu (control E. coli DH5α™ transformed with the pBluescript SK vector without any insert). However, the cell density of E. coli DH5α™ was similar. The cell density of E. coli DH5α™ with pPos was 37.67 x 107,93.33 x 107 CFU·ml-1 at 24 h and 48 h respectively, and the cell density of E. coli DH5α™ with pBlu was 21.33 x 107,70.67 x 107 CFU·ml-1 at 24 h and 48 h respectively.
12We tested whether the decrease in pH in the presence of E. coli was correlated with organic acid secretion. Escherichia coli DH5α™ (pPos and pBlu) secreted acetic acid, and the acetic acid concentration was 168.6 mg·l-1 after 48 h. However, the acetic acid concentration reached 567.3 mg·l-1 by E. coli DH5α™ (pPos), which was significantly higher than those from E. coli DH5α™ (pBlu) (Figure 2b).
3.3. Substrate utilization
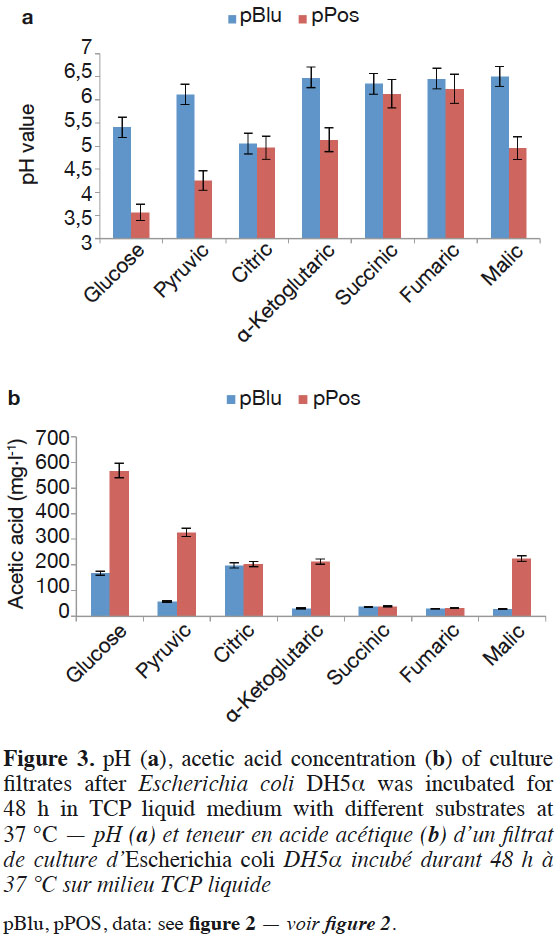
13Glucose, pyruvic acid, citric acid, α-ketoglutaric acid, succinic acid, fumaric acid, and malic acid participate in the tricarboxylic acid cycle of E. coli DH5α™. Escherichia coli DH5α™ containing the target gene was grown in TCP liquid medium with one of the six organic acids as sole carbon sources instead of glucose to demonstrate the function of the cloned pPos sequence. The pH value significantly decreased and the acetic acid concentration increased with E. coli DH5α™ bearing pPos compared to E. coli DH5α™ (pBlu) in the TCP liquid medium after 48 h with glucose, pyruvic acid, α-ketoglutaric acid, and malic acid. The acetic acid concentration reached 567.3, 326.4, 213.2, and 225.3 mg·l-1, respectively. No difference was found in the pH values and acetic acid contents between E. coli DH5α™ (pPos) and E. coli DH5α™ (pBlu) in TCP liquid medium with citric acid, succinic acid, and fumaric acid (Figure 3a and Figure 3b). The hypothetical protein could improve glucose, pyruvic acid, α-ketoglutaric acid, and malic acid utilization in E. coli.
4. Discussion
14In this study, P. oxalicum I1 secreted a high amount of oxalic acid that reached 593.9 μg·ml-1 at 30 h in culture, which decreased the pH of culture from 6.90 to 1.65, and showed a strong ability to convert a wide range of insoluble phosphate into soluble forms (Gong et al., 2014b). Genes that can induce secretion of organic acids are present in the P. oxalicum I1 genome. Therefore P. oxalicum I1 was as a suitable organism from which to clone gene that may enhance organic acids secretion. From P. oxalicum I1, a total of 106clones were obtained as candidates, and the cDNA sequence I-2 we obtained showed only 68% identity and 73% query cover with fungi. The deduced amino acid sequence showed 100% identity with a hypothetical protein in P. oxalicum 114-2, for which no gene function had been reported. The protein sequence from P. oxalicum I1 could be aligned with another hypothetical protein of the Penicillium genus. Even though for some alignments the identity is less than 63%, it clearly forms a group of proteins that most probably share the same function. In this study, we proved that the protein can improve acetic acid secretion in E. coli DH5α™. In addition, the E. coli DH5α™ containing the target gene secreted up to 567 mg·l-1 acetic acid within 48 h. Lü et al. (2012) cloned a full-length gene encoding mitochondrial malate dehydrogenase from P. oxalicum, which was expressed in E. coli to secrete malic, lactic, acetic, and citric acids, and the obtained amount of organic acids was < 60 mg·l-1. Gong et al. (2014a) cloned the delta-1-pyrroline-5-carboxylate dehydrogenase gene that secreted acetic acid and α-ketoglutaric acids, and the obtained the acetic acid concentration was 389.81 μg·ml-1.
15Many organic acid metabolic pathways, such as glycometabolism and tricarboxylic acid cycle, are also present in E. coli. Glucose, pyruvic acid, citric acid, α-ketoglutaric acid, succinic acid, fumaric acid, and malic acid are the main substrates in the tricarboxylic acid cycle of E. coli DH5α™. Escherichia coli DH5α™ containing the target gene showed improved acetic acid secretion with the use of glucose, pyruvic acid, α-ketoglutaric acid, and malic acid. By contrast, acetic acid secretion was not improved when citric acid, succinic acid, and fumaric acid were used. This result indicated that the hypothetical protein was related to the utilization of glucose, pyruvic acid, α-ketoglutaric acid, and malic acid in E. coli.
5. Conclusions
16We obtained a novel gene from Penicillium oxalicum I1 whose overexpression in E. coli DH5α™ increased the secretion of acetic acid. Alignment of the putative amino acid sequence of the protein with other comparable sequences of the Penicillium genus suggests that this is a protein group sharing similar functions, nothing however is known about their role in the other organisms. The results reported here may help to understand the function of these genes and the corresponding protein group.
17Acknowledgements
18Project 41440008 supported by National Natural Science Foundation of China.
Bibliographie
Altomare C., Norvell A., Björkman T. & Harman G., 1999. Solubilization of phosphates and micronutrients by the plant growth promoting and biocontrol fungus Trichoderma harzianum Fifai 1295-22. Appl. Environ. Microbiol., 65, 2926-2933.
Banik S. & Dey B., 1982. Available phosphate content of an alluvial soil as influenced by inoculation of some isolated phosphate-solubilizing micro-organisms. Plant Soil, 69, 353-364.
Bianco C. & Defez R., 2010. Improvement of phosphate solubilization and medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl. Environ. Microbiol., 76, 4626-4632.
Dessureault-Rompré J., Nowack B., Schulin R. & Luster J., 2007. Spatial and temporal variation in organic acid anion exudation and nutrient anion uptake in the rhizosphere of Lupinus albus L. Plant Soil, 301, 123-134.
Fomina M. et al., 2005. Role of oxalic acid overexcretion in transformations of toxic metal minerals by Beauveria caledonica. Appl. Environ. Microbiol., 71, 371-381.
Goldstein A., Braverman K. & Osorio N., 1999. Evidence for mutualism between a plant growing in a phosphate-limited desert environment and a mineral phosphate solubilizing (MPS) rhizobacterium. FEMS Microbiol. Ecol., 30, 295-300.
Gong M., Tang C. & Zhu C., 2014a. Cloning and expression of delta-1-pyrroline-5-carboxylate dehydrogenase in Escherichia coli DH5α improves phosphate solubilization. Can. J. Microbiol., 60(11), 761-765.
Gong M., Du P., Liu X. & Zhu C., 2014b. Transformation of inorganic P fractions of soil and plant growth promotion by phosphate-solubilizing ability of Penicillium oxalicum I1. J. Microbiol., 52, 1012-1019.
Gulati A. et al., 2010. Organic acid production and plant growth promotion as a function of phosphate solubilization by Acinetobacter rhizosphaerae strain BIHB 723 isolated from the cold deserts of the trans-Himalayas. Arch. Microbiol., 192, 975-983.
Khan M., Zaidi A. & Wani P., 2007. Role of phosphate-solubilizing microorganisms in sustainable agriculture - A review. Agron. Sustain. Dev., 27, 29-43.
Kim C. et al., 2003. Cloning and expression of pyrroloquinoline quinine (PQQ) genes from a phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol., 47, 457-461.
Kim K., Jordan D. & Krishnan H., 1998. Expression of genes from Rahmella aquatilis that are necessary for mineral phosphate solubilization in Escherichia coli. FEMS Microbiol. Lett., 159, 121-127.
Lü J., Gao X., Dong Z. & An L., 2012. Expression of mitochondrial malate dehydrogenase in Escherichia coli improves phosphate solubilization. Ann. Microbiol., 62, 607-614.
Meulenberg J. et al., 1992. Nucleotide sequence and structure of the Klebsiella pneumonia pqq operon. Mol. Gen. Genet., 232, 284-294.
Nautiyal C., 1999. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett., 182, 265-270.
Singh K., ArchnaSuman P. & SinghMenhi La., 2007. Yield and soil nutrient balance of a sugarcane plant–ratoon system with conventional and organic nutrient management in sub-tropical India. Nutr. Cycl. Agroecosyst., 79, 209-219.
Walpola B. &Yoon M., 2013. Phosphate solubilizing bacteria: assessment of their effect on growth promotion and phosphorous uptake of mung bean (Vigna radiata (L.) R. Wilczek). Chil. J. Agric. Res., 73, 275-281.






