- Home
- Volume 17 (2013)
- numéro 4
- Role of satellite RNAs in cucumber mosaic virus-host plant interactions: a review
View(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Role of satellite RNAs in cucumber mosaic virus-host plant interactions: a review

Editor's Notes
Received on February 27, 2013; accepted on July 30, 2013
Résumé
Synthèse bibliographique : rôle de l’ARN satellite dans les interactions plantes-virus de la mosaïque du concombre. Un ARN subviral sans fonction codante désigné ARN satellite (ARNsat) est souvent associé au virus de la mosaïque du concombre (CMV). Cet ARNsat a besoin de son virus assistant (dans ce cas précis, le CMV) pour sa réplication et son encapsidation et peut, dans certains cas, modifier les symptômes causés par le CMV soit en les atténuant, soit en les aggravant. Plusieurs études ont montré que cet effet pourrait être dû à une compétition entre l’ARNsat et le CMV pour la réplication. Parallèlement, certains auteurs ont révélé que cette pathogénicité de l’ARNsat serait liée à des séquences primaires spécifiques ou à sa structure secondaire, bien que cet effet soit aussi fonction de la plante hôte infectée et de la souche du virus assistant. Cependant, tout récemment, il a été montré que la réplication des ARNsat était associée à la production de petits ARNs d’environ 21-25 nucléotides dérivant de ces ARNsat et connus pour jouer un rôle dans le phénomène silencing de l'ARN, un mécanisme de défense de la plante primordial dans l’extinction des gènes viraux. Cette revue fait le point sur les connaissances actuelles et la compréhension des mécanismes mis en jeu par les ARNsat pour moduler l’expression des symptômes chez les plantes hôtes infectées par le CMV.
Abstract
Subviral non-coding RNA molecules, known as satellite RNAs (satRNAs), are often associated with cucumber mosaic virus (CMV). These satRNAs require a helper virus (CMV in this case) for their replication, encapsidation and transmission. They modify CMV pathogenicity by either attenuating disease symptoms or by exacerbating them. This effect could be due either to competition between a helper virus and satRNAs for replication, or to specific satRNA sequences or secondary structures. The type of host plant and the CMV strain also affect the behavior of satRNAs. Recent studies have shown that satRNA replication is associated with the production of satRNA-derived small RNAs of 21-25 nucleotides in length, which play a key role in RNA silencing and could explain differences in CMV symptom severity. This review highlights the current understanding and recent advances in relation to satRNA-mediated disease symptoms in CMV-infected plants.
Table of content
1. Introduction
1Cucumber mosaic virus (CMV), the type member of the genus Cucumovirus in the family Bromoviridae, has isometric particles of approximately 29 nm in diameter (Palukaitis et al., 2003). CMV is one of the most widespread plant viruses in the world and it has an extensive host range. CMV infects about 1,200 plant species and more than 75 aphid species are capable of transmitting the virus in a non-persistent manner (Palukaitis et al., 1992; Palukaitis et al., 2003). Numerous CMV strains have been described and classified into two major subgroups based on serological relationships and nucleotide sequence similarities: CMV I and CMV II (Palukaitis et al., 1992). Following phylogenetic analyses, subgroup I was further divided into groups IA and IB (Roossinck et al., 1999). CMV has a tripartite genome of positive-sense single-stranded RNAs, termed RNA1, RNA2 and RNA3. It also contains subgenomic RNAs, designated RNA4 and RNA4A (Palukaitis et al., 2003). RNA1 and RNA2 encode proteins required for CMV replication and virulence (proteins 1a and 2a, respectively) (Kang et al., 2012; Mochizuki et al., 2012). RNA2 also encodes protein 2b, which is translated from subgenomic RNA4A (Du Z.Y. et al., 2007; Mochizuki et al., 2012). Protein 2b also acts as a viral suppressor of RNA silencing and plays a role in virus transport (Zhang et al., 2006; Du Z.Y. et al., 2007; Mochizuki et al., 2012). RNA3 encodes both a protein involved in movement (MP) and a coat protein (CP). This CP is translated from subgenomic RNA4 and plays a role in cell-to-cell and systemic movement. It has also been implicated in symptom modulation but it is not required for virus replication (Mochizuki et al., 2012).
2In addition to genomic and subgenomic RNAs, some CMV strains encapsidate subviral RNAs known as satellite RNAs (satRNAs), which differ from the CMV genome by being dispensable for normal CMV replication (Simon et al., 2004). Satellite RNAs are linear RNA molecules of 332 to 405 nucleotides (nt) without any apparent functional open reading frames (ORFs) (Palukaitis et al., 2003). SatRNA replication is totally dependent on viral replicase activity (Gal-On et al., 1995). SatRNAs can have different effects on CMV replication, pathogenesis, and symptom expression, depending on the host plant and the CMV strain (Garcia-Arenal et al., 1999).
3Viral infections generally cause disease symptoms by interfering with the host plant’s metabolism. Several studies have demonstrated that RNA silencing in plants limits the accumulation of foreign RNA species (Qu, 2010; Zhu et al., 2012). The first biological function established for RNA silencing is the production of small RNAs that are 21-25 nucleotides long and can regulate gene expression in a sequence-specific manner (Zhu et al., 2012). RNA silencing can reduce the expression of specific genes through post-transcriptional and transcriptional gene silencing (Zhu et al., 2012). Post-transcriptional RNA silencing is mediated by short interfering RNAs (siRNAs) and by microRNAs (miRNAs) (Jones-Rhoades et al., 2006; Mallory et al., 2006). Several characteristics are distinct between the two small RNA classes, although the main fundamental difference is the nature of their precursor: siRNAs are processed from long, double-stranded (ds)RNAs, whereas miRNAs are encoded in the plant genome and processed from single-stranded RNA molecules that include an imperfect stem-loop secondary structure (Jones-Rhoades et al., 2006; Qu, 2010). In order to counter this defense mechanism, most plant viruses encode for suppressors of host RNA silencing (Burgyan et al., 2011). SatRNAs associated with helper viruses are also resistant to RNA silencing-mediated degradation, thus suggesting a possible role of this mechanism in the pathogenicity of these subviral RNAs (Wang et al., 2004).
4In this review, the role of satRNAs in CMV-host plant interactions is examined in the light of recent advances in research. Major emphasis is placed on the effect of satRNAs on the components of CMV helper viruses and their pathogenicity, along with the potential interference between satRNAs and RNA silencing.
2. Origin and diversity of satRNAs
5The origin of satRNAs remains enigmatic and subject to much speculation (Roossinck et al., 1992; Simon et al., 2004). To date, no significant sequence identity has been reported between satRNAs and the CMV helper virus and no complete satRNA sequence has been found in any plant genome (Simon et al., 2004; Hajimorad et al., 2009). Several studies show that repeated infections by naturally occurring CMV strains under experimental conditions may occasionally result in the “spontaneous” emergence of satRNAs (Palukaitis et al., 1992; Roossinck et al., 1992). By contrast, other studies involving eleven serial transfers of CMV RNA transcripts on Nicotiana tabacum cv. ‘Ky 14’ did not result in emergence of any satRNAs, whereas a satRNA was detected after at least eight successive transfers of other CMV strains under the same conditions (Hajimorad et al., 2009). Furthermore, certain CMV strains were found unable to support the replication of satRNAs in any of the host plants tested (McGarvey et al., 1995; Yamaguchi et al., 2005).
6More than 100 satRNAs associated with CMV isolates originating from geographically different countries and host plants, of which most were Solanaceae plants such as tomato and tobacco, have been characterized by sequencing and nucleotide sequences were deposited in the National Center for Biotechnology Information (NCBI) GenBank database. These CMV isolates can be classified into at least three groups depending on whether the disease symptoms are unaffected, exacerbated or attenuated by the presence of satRNA on tomato indicator plants (Collmer et al., 1992; Garcia-Arenal et al., 1999). CMV satRNAs have been found in association with both CMV subgroups I and II (Atencio et al., 1997; Mavrodieva et al., 1998).
3. CMV satRNA sequences and secondary structures involved in symptom expression
7CMV satRNAs can differently affect symptom expression depending on the host plant. For example, D satellite RNA (D-satRNA) induces necrosis in infected tomato plants in the presence of CMV but attenuates symptoms in tobacco (Garcia-Arenal et al., 1999). Likewise, CMV-PepY-satRNA isolated from pepper induces chlorosis in hot pepper, but not in tobacco or tomato (Choi et al., 2011). Strain K8-satRNA attenuates symptoms in tobacco and tomato but has no effect on symptoms in squash. In some cases, symptom expression can change in the same host. A CMV satRNA that causes very mild symptoms on many plant species causes more severe symptoms on several hosts after serial inoculations (Alvarez et al., 2003).
8Several authors hypothesized the action of specific satRNA sequences, the competitiveness of helper virus components, and the misregulation of host plant factors in symptom expression. Many satRNAs inducing necrosis, chlorosis or bright yellow mosaic as well as satRNAs carrying disease attenuation domains were identified (Collmer et al., 1992; Garcia-Arenal et al., 1999; Palukaitis et al., 2003; Choi et al., 2011). However, the link between chlorosis symptoms and the chlorosis-domain is not obvious because in some cases satRNA symptom can occur either in tobacco or in tomato, but not in both plant species (Garcia-Arenal et al., 1999).
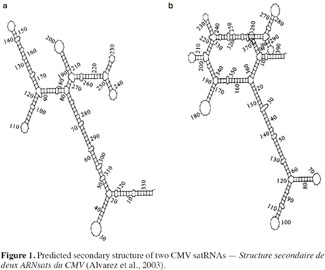
9Several authors have studied the sequences of satRNA variants that confer the ability to induce necrosis on tomato (Devic et al., 1990; Sleat et al., 1994). Necrosis induction has been shown to depend on a domain that maps within the 3′ half of satRNAs, while sequences outside this domain could also influence necrosis extension (Palukaitis et al., 2003). The necrogenic satRNA variants for tomato contain the “consensus” sequence: GA-GCUAAGGCUUA---UGCUAUGCUGAU (Devic et al., 1990). This type of satRNA, which induces systemic necrosis in tomato plants, does not usually cause similar symptoms in other plant species (Garcia-Arenal et al., 1999; Betancourt et al., 2011). In fact, studies of satRNAs nucleotide sequences failed to fully explain the mechanism involved in satRNA pathogenicity. In addition, Y-satRNA has been reported as necrogenic when associated with CMV strain Y (CMV-Y) but not with CMV strain O (CMV-O) and the occurrence of necrosis also depends on the tomato cultivar (Collmer et al., 1992). As there is no functional ORF in satRNAs, their biological properties might be related to their secondary structure (Figure 1), among other factors (Garcia-Arenal et al., 1999; Alvarez et al., 2003). Several studies effectively predict that CMV satRNA is expected to have a high degree of secondary structure, with about 50% base pairing (Rodriguez-Alvarado et al., 1997). Nevertheless, in some cases, primary and secondary structure alterations of some CMV satRNAs do not induce differences in pathogenicity (Garcia-Arenal et al., 1999). The effects of satRNAs on their host plants might be related not only to satRNA sequences but also to interactions with the helper strain and host factors (Betancourt et al., 2011).
4. Effect of satRNAs on the accumulation and pathogenicity of CMV
10The presence of CMV-associated satRNAs usually reduces the titer of the helper virus (Escriu et al., 2000; Palukaitis et al., 2003; Liao et al., 2007). This reduction depends on the CMV isolate and the host plant (Jacquemond et al., 1982; Cillo et al., 2007). Some reports suggest that the effect of satRNA could be related to the competitiveness for replication between the helper virus and satRNA (Wu et al., 1995). Gal-On et al. (1995) revealed that in tobacco and zucchini squash plants, satRNA affects the levels of RNA1, and to a lesser extent of RNA2, and of their encoded proteins, that are components of the CMV replicase. In other studies, satRNAs reduced the expression levels of the coat protein gene (Shang et al., 2009).
11The attenuation of symptoms observed in CMV-infected plants was suspected to result from low accumulation of CMV genomic RNAs (Liao et al., 2007). Nevertheless the reduced CMV accumulation in the presence of satRNA is not always associated with satRNA-mediated symptom attenuation since satRNAs that induce necrosis in tomato plants also reduce CMV accumulation (Escriu et al., 2000). Furthermore, satRNAs that exacerbate symptoms caused a higher reduction of CMV accumulation levels than satRNAs that attenuated them (Escriu et al., 2000; Cillo et al., 2007; Feng et al., 2012).
12Several authors investigated the effect of satRNAs on CMV genomic RNAs and especially on the expression of the protein 2b. Liao et al. (2007) and Chen (2010) showed that satRNA lowered CMV RNA accumulation, leading to a significant decrease in protein 2b, and that deleting the 2b gene had the same effect on genomic RNA replication and/or accumulation as the addition of satRNAs. As these authors, Hou et al. (2011) found that SD-satRNA attenuated the yellowing phenotype induced by SD-CMV in infected Nicotiana benthamiana and Arabidopsis plants in correlation with low accumulation levels of 2b-coding subgenomic RNA4A.
13SatRNAs could serve as a target for RNA silencing, thus playing a role in protecting helper RNAs, especially subgenomic RNA4A (Hou et al., 2011). Wang et al. (2004) succeeded in reducing the symptoms caused by the Y satellite of CMV using a silencing suppressor that prevents RNA silencing in tobacco. These data provided evidence that silencing is probably involved in the pathogenicity of satRNAs, as demonstrated for viroids (Wang et al., 2004).
5. Effects of CMV satRNAs on RNA silencing
14SatRNAs interfere with the pathogenicity of helper viruses and the RNA-silencing pathways of host plants even though RNA silencing also plays a role in the pathogenicity of satRNAs (Wang et al., 2004). There is a limited number of reports on the pathways used by satRNAs to interfere with RNA silencing. Nevertheless, siRNA and miRNA pathways appear to be involved (Zhu et al., 2011; Feng et al., 2012).
15In plants, RNA silencing involving gene suppression requires key components such as Argonaute proteins (AGOs), Dicer or Dicer-like (DCLs) and RNA-directed RNA polymerases (RDRs) (Jones-Rhoades et al., 2006). The CMV protein 2b has been demonstrated to either inhibit the production of RDRs (RDR1 and RDR6) dependant viral siRNAs (Diaz-Pendon et al., 2007; Wang et al., 2010) or inhibit the cleavage activity of AGO1 (Zhang et al., 2006).
16Hou et al. (2011) showed that satRNAs are targeted by RNA silencing, resulting in reduced CMV RNA-derived siRNA production. Furthermore, lower levels of CMV-derived siRNAs are detected in plants infected with SD-satRNA in the presence as well as in the absence of the 2b protein, whereas abundant SD-satRNA-derived siRNAs are detected in the same conditions. Previous works indicate that DCL4, an RNase III-like enzyme, was the primary producer of SD-satRNA-derived siRNAs (Du Q.S. et al., 2007). Likewise, a virus-derived siRNA targeting viral RNAs triggers the host RDR6-mediated degradation of viral RNAs (Zhu et al., 2011). Furthermore, satRNA-derived siRNAs could be associated with different AGO proteins in host plants (Zhu et al., 2011). In fact, satRNA-derived siRNAs could play an important biological function in interactions between hosts, viruses, and satRNAs to determine the final outcome of viral infection (Zhu et al., 2011). In some cases, the production of satRNA-derived siRNAs could lead to attenuated CMV-induced symptoms (Zhu et al., 2011).
17In order to assess the effect of satRNAs on RNA silencing, Feng et al. (2012) analyzed the interference of a benign satRNA (satRNAYn12) and an aggressive satRNA (satRNAT1) with the miRNA-mediated regulation of gene expression in tomato. Infection by CMV-Fny and CMV-Fny-satRNAT1 significantly altered the normal miRNA-mediated gene expression regulation in host plants whereas the ability of CMV-Fny to interfere with miRNA pathways was dramatically reduced when satYn12 was added (Feng et al., 2012). These results showed a differential effect of two distinct satRNAs on miRNA-regulated gene expression in tomato. On one hand, the complex mechanism whereby satRNAs participate in CMV-tomato interactions suggests that disease symptom severity is to some extent positively correlated with interference of miRNA pathways in tomato but, on the other hand, attenuated disease symptoms can be attributed to satRNAs reducing interference of protein 2b with miRNA (Cillo et al., 2009; Feng et al., 2012) or siRNA (Zhu et al., 2011) pathways.
18To further analyze the mechanisms of satRNA-mediated symptom expression, research teams from two different laboratories demonstrated that the sequence complementarity between CMV-Y-satRNA (Y-satRNA) and a chlorophyll biosynthetic gene (Chll) was essential for inducing yellow symptoms on tobacco or pepper (Shimura et al., 2011; Smith et al., 2011). These authors demonstrated that the yellowing symptoms were a result of Y-satRNA siRNA-mediated RNA silencing of the Chll gene, thus leading to the inhibition of chlorophyll biosynthesis (Shimura et al., 2011; Smith et al., 2011). Previous studies revealed that the same yellow phenotype was observed when the Chll genes of tobacco or cotton were targeted by virus-induced gene silencing (Petersen et al., 2005; Tuttle et al., 2008). The Chll gene of pepper and tobacco but not of tomato and Arabidopsis thaliana has a nucleotide sequence stretch that is complementary to the Y-satRNA yellow region (YR) sequence but differs between large amounts of siRNAs produced from the Y-satRNA YR sequence accumulate in Y-satRNA-infected plants (Shimura et al., 2011). The authors proposed a scenario in which AGO1, associated with the primary Y-satRNA-derived siRNAs, cleaves the Chll mRNA at the Y-satRNA YR portion. This cleavage leads to the production of Chll dsRNA by the host RDR, which are subsequently processed into secondary siRNAs (Shimura et al., 2011). These findings shed light on how satRNA-derived siRNAs modulate viral disease symptoms through RNA silencing-based regulation of host genes.
19Other studies showed that satRNA accumulation does not correlate with RNA silencing (Cillo et al., 2004). Xu et al. (2003) studied the necrosis induced by CMV-D-satRNA in tomato and demonstrated the involvement of programmed cell death (PCD) in symptom expression. Previous studies also showed that D-satRNA causes nuclear DNA fragmentation and chromatin condensation in necrotic tissues (Xu et al., 2000). Furthermore, Xu et al. (2000) found that D-satRNA localization in tissue, vascular cell development and induction of spatial patterns of necrosis were correlated.
6. Conclusion
20Advancing our understanding of the role of satRNAs in CMV-plant interactions remains fundamental. Recent studies indicate that CMV-satRNAs can be the target of RNA silencing. SatRNAs can reduce the expression level of the 2b suppressor protein and consequently interfere with CMV pathogenicity. Nevertheless, interaction among satRNA, CMV and host during RNA silencing is not entirely elucidated although a state of relative balance is needed so that none of the three players is eliminated (Zhu et al., 2012). The genetic structure and dynamics of CMV populations and their satRNAs are unrelated (Alonso-Prados et al., 1998), although CMV could play an important role in the selection of specific satRNA variants from mixed populations (Moriones et al., 1991). Likewise, satRNA-induced symptoms can vary. Further studies on CMV, satRNAs and host RNA silencing will lead to a better understanding of interactions among the three players of this important pathosystem.
21Acknowledgements
22We would like to thank Professor Chikara Masuta (Research Faculty of Agriculture, Hokkaido University, Japan) and Professor Marc Fuchs (Cornell University, United States) for their comments and suggestions for improving this manuscript.
Bibliographie
Alonso-Prados J.L. et al., 1998. Satellite RNA of cucumber mosaic cucumovirus spreads epidemically in natural populations of its helper virus. Phytopathology, 88, 520-524.
Alvarez S.P., Xue C.-Y. & Zhou X.-P., 2003. Emergence of new satellite RNA from cucumber mosaic virus isolate P1. J. Zhejiang Univ. Sci., 4, 336-339.
Atencio F.A. et al., 1997. Detection of both subgroups I and II of cucumber mosaic cucumovirus and their satellite RNAs on pepper in Argentina. Plant Dis., 81, 695.
Betancourt M., Fraile A. & Garcia-Arenal F., 2011. Cucumber mosaic virus satellite RNAs that induce similar symptoms in melon plants show large differences in fitness. J. Gen. Virol., 92, 1930-1938.
Burgyan J. & Havelda Z., 2011. Viral suppressors of RNA silencing. Trends Plant Sci., 16, 265-272.
Chen J., 2010. Gene function of cucumber mosaic virus and its satellite RNA regarding viral-host interactions. In: Chen J., ed. Experimental plant virology (Advanced topics in science and technology in China, 1). Hangzhou, China: Zhejiang University Press, 125-162.
Choi S.-K., Jeon Y.W., Yoon J.-Y. & Choi J.-K., 2011. Characterization of a satellite RNA of cucumber mosaic virus that induces chlorosis in Capsicum annuum. Virus Genes, 43, 111-119.
Cillo F. et al., 2004. Analysis of mechanisms involved in the cucumber mosaic virus satellite RNA-mediated transgenic resistance in tomato plants. Mol. Plant-Microbe Interact., 17, 98-108.
Cillo F. et al., 2007. Response of tomato and its wild relatives in the genus Solanum to cucumber mosaic virus and satellite RNA combinations. J. Gen. Virol., 88, 3166-3176.
Cillo F., Mascia T., Pasciuto M.M. & Gallitelli D., 2009. Differential effects of mild and severe cucumber mosaic virus strains in the pertubation of microRNA-regulated gene expression in tomato map to the 3’ sequence of RNA 2. Mol. Plant-Microbe Interact., 22, 1239-1249.
Collmer C.W. & Howell S.H., 1992. Role of satellite RNA in the expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol., 30, 419-442.
Devic M., Jaegle M. & Baulcombe D., 1990. Cucumber mosaic virus satellite RNA (strain Y): analysis of sequences which affect systemic necrosis on tomato. J. Gen. Virol., 71, 1443-1449.
Diaz-Pendon J.A., Li F., Li W.X. & Ding S.W., 2007. Suppressor of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell, 19, 2053-2063.
Du Q.-S. et al., 2007. DCL4 targets cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol., 81, 9142-9151.
Du Z.-Y. et al., 2007. 2b ORF encoded by subgroup IB strains of cucumber mosaic virus induce differential virulence on Nicotiana species. J. Gen. Virol., 88, 2596-2604.
Escriu F., Perry K.L. & Garcia-Arenal F., 2000. Transmissibility of cucumber mosaic virus by Aphis gossypii correlates with viral accumulation and is affected by the presence of its satellite RNA. Phytopathology, 90, 1068-1072.
Feng J. et al., 2012. Differential effects of Cucumber mosaic virus satellite RNAs in the pertubation of microRNA-regulated gene expression in tomato. Mol. Biol. Rep., 39, 775-784.
Gal-On A., Kaplan I. & Palukaitis P., 1995. Differential effects of satellite RNA on the accumulation of Cucumber mosaic virus RNAs and their encoded proteins in tobacco vs zucchini squash with two strains of CMV helper virus. Virology, 208, 58-66.
Garcia-Arenal F. & Palukaitis P., 1999. Structure and functional relationships of satellite RNAs of cucumber mosaic virus. In: Vogt P.K. & Jackson A.O., eds. Satellites and defective viral RNAs. Berlin: Springer-Verlag Press, 37-63.
Hajimorad M.R., Ghabrial S.A. & Roossinck M.R., 2009. De novo emergence of a novel satellite RNA of cucumber mosaic virus following serial passages of the virus derived from RNA transcripts. Arch. Virol., 154, 137-140.
Hou W.-N. et al., 2011. Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by cucumber mosaic virus infection. Mol. Plant Pathol., 12, 595-605.
Jacquemond M. & Leroux J.-P., 1982. L’ARN satellite du virus de la mosaïque du concombre. II. Étude de la relation virus-ARN satellite chez divers hôtes. Agronomie, 2, 55-62.
Jones-Rhoades M.W., Bartel D.P. & Bartel B., 2006. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol., 57, 19-53.
Kang W.-H. et al., 2012. Helicase domain encoded by cucumber mosaic virus RNA1 determines systemic infection of Cmr1 in Pepper. PloS ONE, 7, e43136.
Liao Q. et al., 2007. Satellite RNA-mediated reduction of cucumber mosaic virus genomic RNAs accumulation in Nicotiana tabacum. Acta Biochim. Biophys. Sin., 39, 217-223.
Mallory A.C. & Vaucheret H., 2006. Functions of microRNAs and related small RNAs in plants. Nat. Genet., 38, S31-S36.
Mavrodieva V.A., Barbara D.J. & Spence N.J., 1998. Subgroup determination of Bulgarian isolates of cucumber mosaic virus and the presence of satellite RNAs. Plant Dis., 82, 960.
McGarvey P. et al., 1995. The complete sequence of a cucumber mosaic virus from Ixora that is deficient in the replication of satellite RNAs. J. Gen. Virol., 76, 2257-2270.
Mochizuki T. & Ohki S.T., 2012. Cucumber mosaic virus: virus genes as virulence determinants. Mol. Plant Pathol., 13, 217-225.
Moriones E., Fraile A. & Garcia-Arenal F., 1991. Host-associated selection of sequence variants from a satellite RNA of cucumber mosaic virus. Virology, 184, 465-468.
Palukaitis P., Roossinck M.J., Dietzgen R.G. & Francki R.I.B., 1992. Cucumber mosaic virus. Adv. Virus Res., 41, 281-348.
Palukaitis P. & Garcia-Arenal F., 2003. Cucumoviruses. Adv. Virus Res., 62, 241-323.
Petersen B.O. & Albrechtsen M., 2005. Evidence implying only unprimed RdRP activity during transitive gene silencing in plants. Plant Mol. Biol., 58, 575-583.
Qu F., 2010. Plant viruses versus RNAi: simple pathogens reveal complex insights on plant antimicrobial defense. WIREs RNA, 1, 22-33.
Rodriguez-Alvarado G. & Roossinck M.J., 1997. Structural analysis of necrogenic strain of cucumber mosaic cucumovirus satellite RNA in planta. Virology, 236, 155-166.
Roossinck M.J., Sleat D. & Palukaitis P., 1992. Satellite RNAs of plant viruses: structures and biological effects. Microbiol. Rev., 56, 265-279.
Roossinck M.J., Zhang L. & Hellwald K.H., 1999. Rearrangements in the 5’ non-translated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol., 73, 6752-6758.
Shang J. et al., 2009. Effect of two satellite RNAs on Nicotiana glutinosa infected with cucumber mosaic virus (CMV). Physiol. Mol. Plant Pathol., 74, 184-190.
Shimura H. et al., 2011. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog., 7, e1002021.
Simon A.E., Roossinck M.J. & Havelda Z., 2004. Plant virus satellite and defective interfering RNAs. New paradigms for a new century. Annu. Rev. Phytopathol., 42, 415-437.
Sleat D.E., Zhang L. & Palukaitis P., 1994. Mapping determinants within cucumber mosaic virus and its satellite RNA for the induction of necrosis in tomato plants. Mol. Plant-Microbe Interact., 7, 189-195.
Smith N.A., Eamens A.L. & Wang M.-B., 2011. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PloS Pathog., 7, e1002022.
Tuttle J.R. et al., 2008. Geminivirus-mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol., 148, 41-50.
Wang M.-B. et al., 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. PNAS, 101, 3275-3280.
Wang X.B. et al., 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 107, 484-489.
Wu G. & Kaper J.M., 1995. Competition of viral and satellite RNAs of cucumber mosaic virus for replication in vitro by viral RNA-dependent RNA polymerase. Res. Virol., 146, 61-67.
Xu P. & Roossinck M.J., 2000. Cucumber mosaic virus D satellite RNA-induced programmed cell death in tomato. Plant Cell, 12, 1079-1092.
Xu P., Blancaflor E.B. & Roossinck M.J., 2003. In spite of induced multiple defense responses, tomato plants infected with cucumber mosaic virus and D satellite RNA succumb to systemic necrosis. Mol. Plant-Microbe Interact., 16, 467-476.
Yamaguchi N. et al., 2005. Genetic mapping of the compatibility between a lily isolate of cucumber mosaic virus and a satellite RNA. J. Gen. Virol., 86, 2359-2369.
Zhang X. et al., 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev., 20, 3255-3268.
Zhu H. et al., 2011. Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3’ untranslated region of cucumber mosaic virus triggers viral RNAs for degradation. J. Virol., 85, 13384-13397.
Zhu H. & Guo H.S., 2012. The role of virus-derived small interfering RNAs in RNA silencing in plants. Sci. China Life Sci., 55, 119-125.
To cite this article
About: Kouakou Théodore Kouadio
Univ. Liege - Gembloux Agro-Bio Tech. Unité de Phytopathologie. Passage des Déportés, 2. B-5030 Gembloux (Belgium). E-mail: tkouadiothed@yahoo.fr – Institut National Polytechnique Félix Houphouet Boigny. Laboratoire de Phytopathologie et de Biologie végétale. Département Agriculture et Ressources Animales. BP 1313. CI-Yamoussoukro (Côte d’Ivoire).
About: Caroline De Clerck
Univ. Liege - Gembloux Agro-Bio Tech. Unité de Phytopathologie. Passage des Déportés, 2. B-5030 Gembloux (Belgium).
About: Thérèse Atcham Agneroh
Institut National Polytechnique Félix Houphouet Boigny. Laboratoire de Phytopathologie et de Biologie végétale. Département Agriculture et Ressources Animales. BP 1313. CI-Yamoussoukro (Côte d’Ivoire).
About: Olivier Parisi
Univ. Liege - Gembloux Agro-Bio Tech. Unité de Phytopathologie. Passage des Déportés, 2. B-5030 Gembloux (Belgium).
About: Philippe Lepoivre
Univ. Liege - Gembloux Agro-Bio Tech. Unité de Phytopathologie. Passage des Déportés, 2. B-5030 Gembloux (Belgium).
About: Haïssam Jijakli
Univ. Liege - Gembloux Agro-Bio Tech. Unité de Phytopathologie. Passage des Déportés, 2. B-5030 Gembloux (Belgium).






