The polyphasic description of a Desmodesmus spp. isolate with the potential of bioactive compounds production
Helwan University. Faculty of Science. Department of Botany and Microbiology. Ain Helwan campus. ET-11795 Helwan (Egypt). E-mail: nerminel_semary@yahoo.co.uk
Received on December 2, 2009, accepted on October 5, 2010
Résumé
Description polyphasique d’un isolat de Desmodesmus spp. présentant un potentiel dans la production de composés bio-actifs. L’approche polyphasique a été utilisée pour décrire une espèce de Desmodesmus formant des colonies. Il s’agit d’un isolat collecté dans le Nil, Aire de Maadi, district de Helwan en Égypte. Cet isolat croît le mieux à température modérée tout en étant exposé à une relativement forte intensité lumineuse. Les caractères phénotypiques révèlent la présence à la fois de cellules et d’agrégats cellulaires comptant de 2 à 4 cellules. Les cellules présentent une largeur maximale de 4-6 µm ± 0,5 pour une longueur de 11-15 µm ± 0,48. Elles sont pourvues de projections épineuses. Les cellules ont un aspect fortement granuleux et elles sont contenues dans un étui mucilagineux commun. Les formes en colonies se développent par la production de cellules filles au sein de la cellule mère. L’analyse moléculaire utilisant le gène 18S rRNA montre une certaine similarité avec Desmodesmus communis qui en est l’organisme le plus proche dans la banque de données. L’analyse phylogénétique regroupe bien cet isolat avec les autres Desmodesmus spp., tout en l’écartant des Scenedesmus spp. de la base de données. La composition en acides gras révèle la présence de l’acide palmitique saturé comme le composant majeur suivi de l’acide palmitoléique (acide gras monoinsaturé), alors que les acides gras polyinsaturés sont peu abondants. L’acide palmitoléique en particulier est supposé être impliqué dans un mécanisme de défense active. Le criblage phytochimique révèle la présence d’alcaloïdes et de saponines ainsi que l’absence de tannins. Les fractions d’extraits méthanoliques montrent des activités antimicrobiennes contre des souches de bactéries pathogènes, y compris celles multirésistantes aux antibiotiques. Cette étude documente la présence de cette souche de Desmodesmus dans le Nil et met en évidence son potentiel biotechnolologique comme source de composés bio-actifs.
Abstract
A polyphasic approach was applied to describe a colony-forming Desmodesmus species collected from the Nile River, Maadi area, Helwan district, Egypt. The isolate grows best at moderate temperature and relatively high light intensity. The phenotypic features revealed the presence of both unicellular and colonial forms of the isolate and the latter form was either 2-4 celled. Cells were 4-6 m ± 0.5 at their widest point and 11-15 m ± 0.48 in their length with spiny projections that encircled the cells. Cells were heavily-granulated and enclosed within common mucilaginous sheath. Colonial forms were developed through production of daughter cells within mother cell. Molecular analysis using 18S rRNA gene showed some similarity to its nearest relative (Desmodesmus communis) whereas the phylogenetic analyses clustered it together with other Desmodesmus spp. and away from Scenedesmus spp. from the database. However, the use of ITS-2 as a phylotaxonomic marker proved to be more resolving and confirmed the generic identity of the isolate as Desmodesmus spp. The fatty acid composition revealed the presence of saturated palmitic fatty acid as the most abundant component followed by monounsaturated palmitoleic acid whereas the polyunsaturated fatty acids were in relatively low abundance. The palmitoleic acid in particular is suggested to be involved in active defense mechanism. The phytochemical screening revealed the presence of alkaloids and saponins and absence of tannins. Fractions of methanolic extracts showed antimicrobial activities against pathogenic bacterial strains including multi-drug resistant ones. This study documents the presence of this strain in the River Nile and highlights its biotechnological potential as a source of bioactive compounds.
1. Introduction
1Several microscopic chlorococcal algae are considered to be of cosmopolitan distribution including some members of the genus Desmodesmus (Coesel et al., 2008). In algal systematics, the taxonomy of Scenedesmus and Desmodesmus genera has been the centre of hot debate for decades as there are hardly clear diagnostic phenotypic features that differentiate between them. According to Wozniak et al. (2008), Scenedesmus was originally described as freshwater, non-motile green-alga in 1829 by Meyen. This description outlined the colony-forming habit of that genus and indicated the possibility of presence of spiny and non-spiny morphological forms. For long, this was largely accepted until Trainor et al. (1976) proposed that spiny and non-spiny forms were in fact two distinct genera according to their different phenotypic and physiological characters. Additionally, An et al. (1999) used ITS-2 region as a molecular marker that supported the separation of Desmodesmus, as a spiny taxon as a spiny taxon. Most of the Desmodesmus species were found to have one or several spines on the cells (Hegewald, 2000; Johnson et al., 2007) whereas those of Scenedesmus were regarded as non-spiny. Another characteristic feature of Desmodesmus is the presence of cell wall layers with ornamentations formed by the outermost layer often visible under the light microscope as granulations (Hegewald, 1978; An et al., 1999). Based on those characteristic features as well as phylogenies based on 18S and ITS-2 rDNA, Desmodesmus was given the taxonomic rank of a genus (Kessler et al., 1997; An et al., 1999; Van Hannen et al., 2002). Recently, Wozniak et al. (2008) suggested that several methodologies should be combined to provide a holistic understanding of these taxa. Despite the ubiquity of this genus worldwide and its biotechnological potentials in heavy metal biosorption (Monterio Cristina et al., 2010) and allelochemical production (Leflaive et al., 2008), the literature lacks a thorough record of the presence and activity of this genus in the River Nile (Vanormelingen et al., 2007). Some of the few records available on the presence of this genus in Egypt include detecting it in the Sacred Lake inside Karnak Temple (Hamed et al., 2003) and in Lake Nasser (El-Otify et al., 2003). The scarcity of information on the presence of the genus Desmodesmus in River Nile is probably due to the morphological overlap between this genus and Scenedesmus. Identification based solely on morphology can lead to mischaracterization of microalgal taxa that share common morphological features and the extreme plasticity of morphological characters can certainly lead to erroneous interpretations (Trainor et al., 1990; Kessler et al., 1997; Trainor, 1998). Therefore, in the present study we combine growth experiments with several morphological, molecular and biochemical features to accurately identify the organism under study. This polyphasic approach is inevitably needed in systematics especially when dealing with desmids and chlorococcal algae (Coesel et al., 2008). Moreover, we also examine the presence of bioactive compounds and the antimicrobial activity of methanolic extract of that organism to explore opportunities for its biotechnological exploitation and application.
2. Materials and methods
2.1. Isolation, growth conditions and culture establishment
2Samples were collected in triplicates, from the photic zone of the banks of The Nile, Maadi area, in sterile containers. Sampling took place in April 2006 at temperature 35°C. Initial microscopic examination (Bosch and Lomb, USA) showed the dominance of green algae and diatoms with few observations of cyanobacteria and chrysophytes. Samples were spun down, and the pellets were spread over different solidified media for green algae including ASM medium (Gorham et al., 1964) and modified Bold's medium (Nichols, 1973). Colonies were picked up, examined under microscope and used to establish monospecific cultures through successive culturing and purification steps. Bold's growth medium gave the best growth recording after modifying its content by adding nicotinamide (Sigma) at different concentrations of which the highest gave highest growth recording. Cultures were also placed under different light / temperature conditions and their optimal growth was recorded at high temperatures ranging from 30-40ºC and high light intensities i.e. 60-100 µmole.m-2.s-1 (12 : 12 L/D). Great growth inhibition was observed at low temperatures below 15°C and low light intensities.
2.2. Phenotypic characters
3Light microscopy. Cell dimensions were determined by light microscopy using a microscope digital camera (3.34 x 106 pixels); Q-imaging digital camera (Micropublisher 5.0 RTV) and Q capture (Quantitative Imaging Corporation) supplied with Image analysis system (Simple PCI 5.3.1, Compix Inc., Cranberry, Pennsylvania, USA). The camera was fitted to a Leitz Orthoplan microscope (Wetzlar, Germany) equipped with a 40 x PHACO and 100 x Oil immersion lenses. Dimensions were taken in ten replicates and pixels were equated to microns using software package. The means and standard deviations of cell dimensions were calculated using statistical package in Excel, Microsoft.
4Scanning electron microscopy. Samples were directly mounted on copper holder and glued onto carbon paste and covered with gold using Ion sputtering (JFC-100E) and examined using JEOL 100-S electron microscope, Japan.
2.3. Molecular analyses
5PCR mixture composition, amplification conditions and sequence deposition. Genomic DNA was extracted using DNA purification kit (Qiagen, The Netherlands). The different genetic loci were initially amplified using SuperTaq polymerase enzyme with SuperTaq buffer (H.T. Biotechnology Ltd, UK) without additional Mg. The primers were used at a final concentration of 0.5 µM in 25-µl reactions, which also contained 0.5 U of Taq polymerase, 200 µM deoxynucleoside triphosphates (dNTPs) (Promega, United Kingdom) and 1x SuperTaq buffer. The reactions contained 2 µl of genomic DNA as a template. Eukaryotic 18S primers were used to initially verify the isolate taxonomic identity: EukF: AACCTGGTTGATCCTGCCAGT and EukR: TGATCCTTCTGCAGGTTCACCTAC designed by Delong (1992). Concerning the ITS-2 region, the primers proposed by Van Hannen et al. (2002) were used; ITS-2 forward (GCAACGATGAAGAACGCAGC) and ITS-2 reverse (CCTCCGCTTATTGATATGC). PCR product was ligated into the TOPO®4.1 vector for sequencing (Invitrogen, The Netherlands). The amplification protocol was 94ºC for 5 min x 1 cycle; 94ºC, 1 min; 55ºC, 1 min; 72ºC, 1.30 min x 35 cycle; 72ºC, 7 min x 1 cycle. The 18S rRNA gene sequence was deposited in the GenBank database under the accession number (EU689108). The sequence retrieved for ITS-2 amplification was deposited at the GenBank under the accession number (FJ178437).
6Phylogenetic reconstruction. For the purpose of phylogenetic analyses, 18S rRNA and ITS-2 genetic sequences from both representative genera Scenedesmus and Desmodesmus were imported from GenBank and aligned in Clustal W tool within alignment function of MEGA 4 Phylogenetic package. Phylogenetic trees were reconstructed using different methods integrated in the MEGA 4 software created by Tamura et al. (2007) such as minimum evolution, maximum parsimony and neighbor-joining using both consensus and linearised tree approaches. Bootstrap values from 500 resamplings were calculated for each set of data and all trees all trees, based on 18S rRNA gene sequence, were rooted using the filamentous cyanobacterium Phormidium uncinatum as an outgroup whereas trees based on ITS-2 sequence were rooted using Scenedesmus jovais. All resulting trees had similar topologies indicating similar phylogenetic relationships. Bootstrapped-consensus maximum parsimony tree was chosen for presentation with representatives from different Desmodesmus and Scenedesmus taxa. Additional single branch analysis was performed to obtain reliable branching. The evolutionary distances were computed using the Maximum Composite Likelihood method. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option).
7Fatty acid analysis. A modified method of Gunasekaran et al. (1980) was applied. Briefly, lyophilized cells were treated with methanolic HCl in the presence of 2, 2-dimethyl propane (used as a hygroscopic substance to allow removal of water from the reaction to facilitate ester bond formation). The reaction proceeded for 2 h at 80ºC followed by addition of 0.9% M NaCl to the reaction mixture. Three hundred microlitres of n-hexane were added and the sample was spun at 3,000 rpm for 5 min. The methyl esters in n-hexane layer were analyzed on a Hewlett-Packard Model 5830A gas chromatography. The flow rate of N2 was 40 ml.min-1 and the glass column, filled with 3% SP-2310/ 2% SP 2300 adsorbed on chromasorb W(80-100 mesh) as stationary phase. Temperature was programmed to increase linearly from 160ºC to 230ºC at 30°C min. The esters were identified by co-chromatography with standards which ranged between C10 - C22: 6 methyl esters of fatty acids at a concentration of 100 µg.ml-1.
8Phytochemical screening. Approximately 1 g of lyophilized sample was extracted in 80% ethanol for two days. The extract was used for the following qualitative tests: test for tannins was carried out according to Claus (1967) where 2 ml of the alcoholic extract was added to 2 ml distilled water and filtered. One ml of 5% ferric chloride was added to the filtrate. The development of a yellowish green color usually indicates tannins presence. Saponins test was performed according to Wall et al. (1954) where they were detected by their ability to develop a froth that is stable for a period of 30 min and longer. Two ml of alcoholic extract were added to 1 ml of distilled water then filtered. The filtrate was vigorously shaken. Test for alkaloid was performed according to Scholz et al. (2006) where the lyophilized sample was boiled in water with 5 ml 2M HCl solution and the filtrate was treated with Mayer’s reagent (1,358 g HgCl2 in 60 ml double distilled water, 5.0 g KI in 10 ml double distilled water, both preparations are mixed together and the total volume completed to 100 ml. The presence of alkaloid compounds is established by the occurrence of turbidity or precipitation.
9Antimicrobial screening
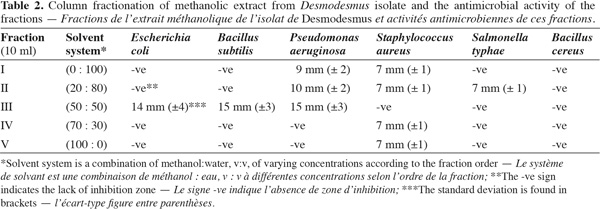
10Extraction and column chromatography of methanolic fractions. Desmodesmus biomass (5 g fresh wt.) was collected and lyophilized. The lyophilized cells (0.5 g dry wt.) were extracted twice with 100 ml methanol HPLC grade for two days, centrifuged (14,000 × g) for 30 min using Hettich-Jenway cooling centrifuge, Germany. The supernatant was left to evaporate to dryness and was dissolved again in methanol (Doan et al., 2000). The sample was applied on a silica gel G60 column (1.5 x 25 cm) prepared from slurry of 30 g of precipitated Silica gel G60 (Merck, UK). The column was developed using the following solvent systems sequence in table 2.
11Antimicrobial bioassay. The multi-drug resistant pathogenic bacterial strains were Escherichia coli and Pseudomonas aeruginosa which were isolated from local clinical samples. Different antibiotics, i.e. streptomycin, vancomycin and rifampicin, were tested using antibiotic discs. Other pathogenic bacterial strains (Helwan Microbial Culture collection) were also tested which included Staphylococcus aureus, Salmonella typhae, Bacillus subtilis and Bacillus cereus. The extracted methanolic fractions were concentrated before applying to 6 mm paper disks (Difco) for testing. The paper was left to dry and evaporate the solvent before using in the antimicrobial test. Sensitivity of these bacterial strains to the extracted fractions was assessed by using the Kirby Bauer Disk Diffusion Susceptibility method (Bauer et al., 1966). The pathogenic bacterial strains were suspended in 5 ml of normal saline solution and the bacterial suspension was then added to 20 ml nutrient agar and poured into Petri dishes after mixing. The plates were incubated for 18 h at 37ºC after which the diameter of the inhibition zone was measured in triplicates and the average values and standard deviation were recorded. Disks containing methanol were left to evaporate and then used as negative controls.
3. Results
3.1. Growth experiments
12The isolate was found to be sensitive to both extremely high (> 45ºC) and low temperatures (< 15ºC) and high continuous illumination. The growth of the isolate responded positively to the increased addition of vitamin B3 and gave best growth, as judged from dry weight records, at temperature range 30-40ºC and relatively high illumination (40 µmol.photon.m-2.s-1). The daughter cells were observed to develop within mother cell during development of colonies.
3.2. Morphological characters
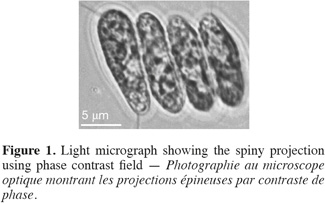
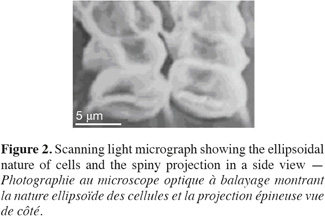
13Cell dimensions were 11-15 µm ± 0.48 in length and 4-6 µm ± 0.5 in its widest point. Cells were either ellipsoidal or obovate and were heavily-granulated (Figure 1). Cells were mostly arranged in tetrads but single and double-celled forms were also observed. The colonial forms always possessed spiny projections. The scanning electron microscopy showed the spiny projections to encircle cells (Figure 2).
3.3. Molecular analyses
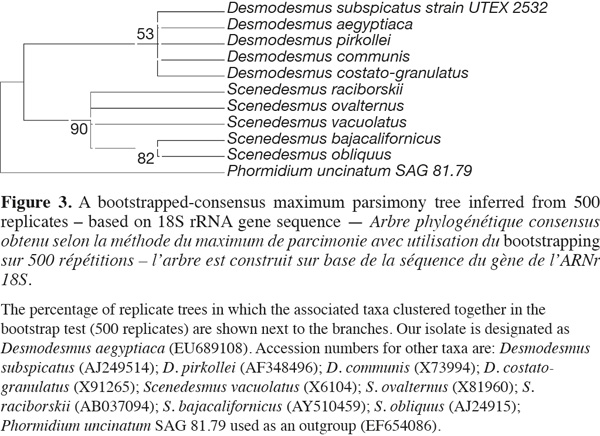
1418S rRNA and phylogenetic reconstruction. The sequence retrieved was compared to other sequences deposited at GenBank using nucleotide BLAST search and showed only 90% similarity with best relative being Desmodesmus communis, but with also closely similar identities to other Desmodesmus and Scenedesmus species. However, the phylogenetic analysis clustered our isolate together with other Desmodesmus spp. and away from Scenedesmus isolates but with low bootstrap values, thus resulting in less reliable phylogenetic inference of the taxonomic identity of that isolate (Figure 3).
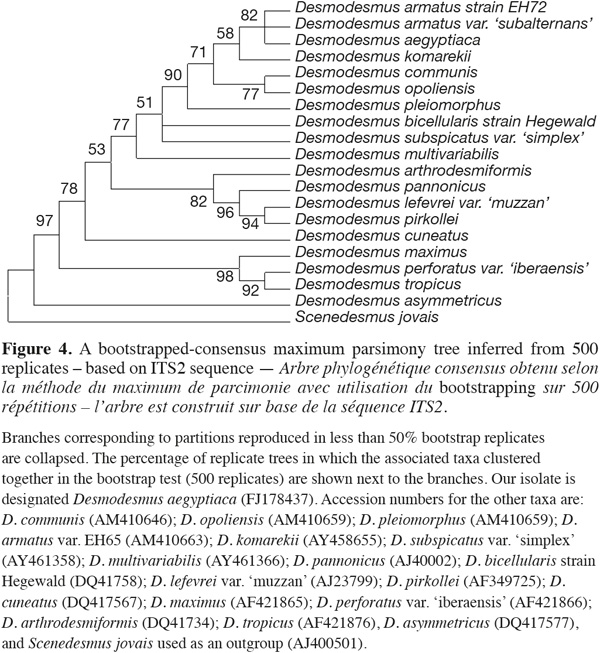
15ITS-2 sequence analysis. The sequence retrieved was compared to other sequences deposited at GenBank using nucleotide BLAST search and showed only 96% similarity with best relative being Desmodesmus armatus but with also closely similar identities to only other Desmodesmus species. The nucleotide sequence of the ITS-2 region of rDNA was analyzed. The ITS-2 secondary structure was predicted using mfold program version 3.1 by Zuker et al. (1999). The phylogenetic analysis and tree reconstruction clustered the Desmodesmus isolate from Egypt, arbitrarily designated as Desmodesmus aegyptiaca, along with other Desmodesmus isolates with high bootstrap values and away from Scenedesmus isolate that was used as an outgroup (Figure 4).
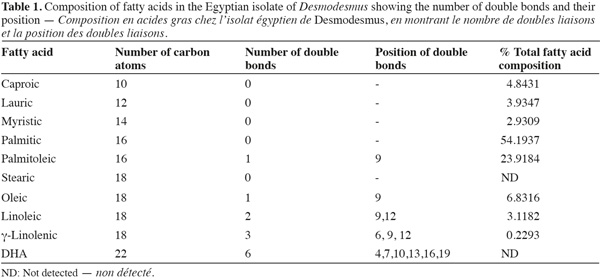
16Fatty acid composition. Fatty acid composition was mostly dominated by saturated fatty acids followed by monounsaturated fatty acids (Table 1). The most abundant fatty acid was palmitic acid (54%) followed by palmitoleic acid (23%). Polyunsaturated fatty acid was a minor component in the fatty acid composition and DHA fatty acid was completely absent.
17Phytochemical screening. The phytochemical screening confirmed the presence of saponins (formation of persistent froth) and alkaloids (the formation of a precipitate). Tannins were absent.
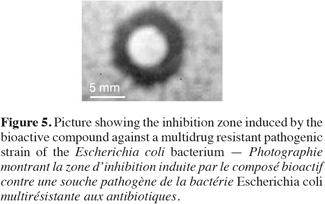
18Antimicrobial bioactivity. Results showed that the multi-drug resistant strains E. coli and P. aeruginosa were highly resistant to all antibiotics tested. Testing the different methanolic fractions against the microbial strains, some bioactivity of the fractions retrieved from column chromatography was observed (Table 2). Fraction III was highest in bioactivity against E. coli (Figure 5), P. aeruginosa and Bacillus subtilis (Table 2). None of the fractions was effective against Bacillus cereus. The disks containing evaporated methanol did not show any inhibition zone.
4. Discussion
19The isolate favored the growth under relatively high temperature and illumination. This was quite expected as the isolate was originally collected from the River Nile which is a subtropical water body. The microalga studied showed characteristic Desmodesmus features. Among those features are the granulated cell wall surface and the spiny projections. The ornamentation of the cell wall is also characteristic of that genus. Recent analyses have suggested that certain morphological characters may well be stable enough for species determinations in Desmodesmus such as the presence of spines and the nature of wall (Hegewald et al., 2005; Leon et al., 2006). In that regard, the presence of spines encircling the cell is an important characteristic of our isolate. However, Vanormelimgen et al. (2007) reported the identification of Desmodesmus species with no spines which only indicates that this morphological character alone is not entirely exclusive for this genus and the characterization of taxa belonging to that genus must be performed in a polyphasic context. In accordance of this, Vanormelingen et al. (2007) reported the lack of studies that combines morphological and molecular data (namely ITS-2 and rDNA phylogeny) together for Desmodesmus species circumscription and indicated the importance of those studies for accurate identification of cryptic isolates.
20The molecular analysis showed that the 18S rRNA genetic marker was not very useful in terms of resolving the specific nature of the isolate under study. Although the isolate was 90% similar to Desmodesmus communis but it was also similar to other Scenedesmus and Desmodesmus species. Therefore, this marker alone cannot resolve the accurate taxonomic designation of that isolate and should be used with the other marker (ITS-2) region as previously suggested by An et al. (1999). Indeed the ITS-2 was more resolving and did show high similarity percentages (up to 96% sequence similarity) to other sequences all derived from Desmodesmus species of different localities. However, the closest relative to our isolate based on sequence of ITS-2 was different from that retrieved based on 18S rRNA and the similarity percentages in both cases was only less than 97% which is strongly indicative that the isolate under study might represent a novel species. In agreement with this, Lewis et al. (2004) used the ITS-2 marker to resolve the identity of cryptic species isolated from desert microbiotic crusts and showed that the ITS-2 sequence differences within each species of Desmodesmus were limited unlike those within Scenedesmus species (Van Hannen et al., 2002). Therefore, new species were erected on the basis of this small substitution difference coupled with habitat differences, despite their morphological similarity.
21The pattern of fatty acids in Chlorococcales (Ahlgren et al., 1992) is mainly characterized by the abundance of palmitic acid (16:0), the presence of considerable amounts of linolenic acid (18:3ω3), linoleic acid 18:2ω9/12 and oleic acid 18:lω9 and the lack of DHA C22:6. Similarly, our results confirmed the presence of palmitic acid in abundance followed by the palmitoleic acid which is not a characteristic fatty acid of chlorophyta. Nevertheless, other chlorophyta-characteristic fatty acids were all found including oleic, linoleic and linolenic fatty acids but the latter accounted only for a minuscule amount of total fatty acids. Saturated fatty acids such as caproic, lauric and myristic were also detected. For organisms living in hot subtropical water bodies, the dominance of total fatty acid composition by saturated fatty acids is an ecological advantage where they effectively strengthen cell membranes under thermophilic conditions as outlined by Madigan et al. (2000). Moreover, palmitoleic acid is suggested to play a role as in the recently discovered “Activated Defence Mechanism”. This fatty acid, along with other mono- and poly-unsaturated fatty acids, are thought to exhibit broad biological functions including toxicity to grazers and inhibitory effects against numerous bacteria (Desbois et al., 2008). Maslova et al. (2004) indicated that this specific fatty acid plays a major role in adaptation to different growth conditions in cyanobacteria.
22The phytochemical screening for saponins and alkaloids were positive. These bioactive compounds have protective functions within algal cells and can be exuded to the outside as a defense mechanism (Scholz et al., 2006). Tannins on the other hand were absent as they are mostly characteristic of brown rather than green algae.
23The general observation that Desmodesmus species is of wide distribution in freshwater bodies world-wide and that cultures of the strain under study grew with no apparent bacterial contamination indicated the potential allelopathic/antimicrobial activity of the isolate. Leflaive et al. (2008) showed that Desmodesmus quadrispira produced inhibitory compounds against Uronem canfervicolum. In our case, the methanolic extract was effective against multi-drug resistant local pathogenic strains. This may imply that this isolate is a promising source of bioactive compounds that may act in active defense against coexisting microbial flora and at the same time its spiny morphological nature may well protect it against grazers.
24Taken all together, the polyphasic approach used in this study provided a holistic description of the morphological, biochemical and molecular features of the isolate. The study also documents the presence of this isolate in the River Nile and implies the possibility that this isolate can be affiliated to a novel Desmodesmus species that is well-adapted to eutrophic subtropical water body and possesses the potential of producing bioactive compounds that are deterrent to other organisms.
25Abbreviations
26ITS: internal transcribed spacer
27DHA: docosahexaenoic acid
Bibliographie
Ahlgren G., Gustafsson I.-B. & Boberg M., 1992. Fatty acid content and chemical composition of freshwater microalgae. J. Phycol., 28, 37-50.
An S.S., Friedl T. & Hegewald E., 1999. Phylogenetic relationships of Scenedesmus and Scenedesmus-like coccoid green algae as inferred from ITS-2 rDNA sequence comparisons. Plant Biol., 1, 418-429.
Bauer A.W., Kirby W.M., Sherris J.C. & Turck M., 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol., 45, 493-496.
Claus E.R., 1967. Pharmacognosy. 5th ed. London: Henry Kimpton Co. Inc.
Coesel P.F.M. & Krienitz L., 2008. Diversity and geographic distribution of desmids and other occoid green algae. Biodivers. Conserv., 17, 381-392.
Delong E.F., 1992. Archaea in coastal marine environments. Proc. Natl Acad. Sci. U.S.A., 89, 5685-5689.
Desbois A.P., Caldwell G.S., Baptie M. & Smith V.J., 2008. Isolation and structural characterization of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol., 81(4),755-764.
Doan N.T., Rickards R.W., Rothschild J.M. & Smith G.D., 2000. Allelopathic actions of the alkaloid 12-epi-Hapalindole E isonitrile and calothrixin A from cyanobacteria of the genera Fischerella and Calothrix. J. Appl. Phycol., 12, 409-416.
El-Otify A.M., Shafik H.M. & Szőke E., 2003. Analyses of physico-chemical characteristics and phytoplankton communities of Lake Nasser during the last two decades. Acta Bot. Hung., 45(1-2), 75-100.
Gorham P.R., Mclachlan J., Hammer U.T. & Kim W.K., 1964. Isolation and culture of toxic strains of Anabaena flos-aquae (Lyngb.) de Brèb. Verh. Int. Ver. Theor. Angew. Limnol., 15, 796-804.
Gunasekaran M. & Hughes W.T., 1980. Gas liquid chromatography, a rapid method for identification of different species of Candida. Mycologia, 72, 505-511.
Hamed A.F., Shafik H.M. & Shaaban A.S., 2003. Phytoplankton and benthic communities of a small water body (Sacred Lake, Karnak Temple) Luxor, Egypt. Acta Bot. Hung., 45(1-2), 101-112.
Hegewald E., 2000. New combinations in the genus Desmodesmus (Chlorophyceae, Scenedesmaceae). Algol. Stud., 96, 1-18.
Hegewald E., Schmidt A., Braband A. & Tsarenko P., 2005. Revision of the Desmodesmus (Sphaeropleales, Scenedesmaceae) species with lateral spines. 2. The multi-spined to spineless taxa. Algol. Stud., 116, 1-38.
Johnson J.L., Fawley M.W. & Fawley K.P., 2007. The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasca State Park, MN, USA. Phycologia, 46(2), 214-229.
Kessler E. et al., 1997. Physiological, biochemical and molecular characters for the taxonomy of the subgenera of Scenedesmus (Chlorococcales, Chlorophyta). Bot. Acta, 110, 244-250.
Leflaive J., Lacroix G., Nicaise Y. & Ten-Hage L., 2008. Colony induction and growth inhibition in Desmodesmus quadrispina (Chlorococcales) by allelochemicals released from the filamentous alga Uronema confervicolum (Ulotrichales). Environ. Microbiol., 10(6), 1536-1546.
Leon S.L. & Hegewald E., 2006. A revision of the species Desmodesmus perforatus and Desmodesmus tropicus (Scenedesmaceae, Chlorophyceae, Chlorophyta). Phycologia, 45, 567-584.
Lewis L.A. & Flechtner V.R., 2004. Cryptic species of Scenedesmus (Chlorophyta) from desert soil communities of western North America. J. Phycol., 40, 1127-1137.
Madigan M.T., Martinko J.M. & Parker J., 2000. Brock biology of microorganisms. London: Prentice-Hall International Ltd.
Maslova I.P. et al., 2004. Lipid fatty acid composition and thermophilicity of cyanobacteria. Russ. J. Plant Physiol., 51(3), 353-360.
Monterio Cristin M., Castro Paula M.L. & Xavier M.F., 2010. Cadmium removal by two strains of Desmodesmus pleiomorphus cells. Water Air Soil Pollut., 208(1-4), 17-27.
Nichols W.H., 1973. Growth media-fresh water. In: Stein J.R., ed. Handbook of phycological methods. Cambridge, UK: Cambridge University Press, 7-14.
Scholz B. & Liebezeit G., 2006. Chemical screening for bioactive substances in culture media of microalgae and cyanobacteria from marine and brackish water habitats: first results. Pharm. Biol., 44 (7), 544-549.
Tamura K., Dudley J., Nei M. & Kumar S., 2007. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol., 24, 1596-1599.
Trainor F.R., 1998. Biological aspects of Scenedesmus (Chlorophyceae): phenotypic plasticity. Nova Hedwigia Beih., 117, 1-367.
Trainor H.R., Cain J. & Shubert L.E., 1976. Morphology and nutrition of the colonial green alga, Scenedesmus: 80 years later. Bot. Rev., 42, 5-25.
Trainor F.R. & Egan P.F., 1990. Phenotypic plasticity in Scenedesmus (Chlorophyta) with special reference to S. armatus unicells. Phycologia, 29, 461-469.
Van Hannen E.J., Fink P. & Lürlig M., 2002. A revised secondary structure model for the internal transcribed spacer 2 of the green algae Scenedesmus and Desmodesmus and its implication for the phylogeny of these algae. Eur. J. Phycol., 37(2), 203-208.
Vanormelimgen P. et al., 2007. The systematics of small spineless Desmodesmus species, D. costato-granulatus (Sphaeropeales, Chlorophyceae), based on ITS 2rDNA sequence analyses and cell wall morphology. J. Phycol., 43(2), 378-396.
Wall M.E. et al., 1954. Steroidal sapogenins. VII-Survey of plants for steroidal sapogenins and other constituents. J. Pharm. Soc., 43, 1-3.
Woźniak E.W. & Shubert E., 2008. Systematics of Desmodesmus and Scenedesmus: a conundrum? In: Proceedings of the 56th meeting of British Phycological Society, January, Bristol, United Kingdom.
Zuker M., Mathews D.H. & Turner D.H., 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J. & Clark B.F.C., eds. Proceedings of the NATO advanced Research Workshop on RNA biochemistry and biotechnology, October 10-17, 1998, Poznan, Poland. Dordrecht, The Netherlands: Kluwer Academic Publishers, 11-43.

