- Portada
- Volume 91 - Année 2022
- Numéro 1
- Optimization of Surface-Assisted Laser Desorption/Ionization Mass Spectrometry
Vista(s): 2125 (43 ULiège)
Descargar(s): 115 (0 ULiège)
Optimization of Surface-Assisted Laser Desorption/Ionization Mass Spectrometry
The quest for the Holy Grail from the study of thermometer ions

Documento adjunto(s)
Version PDF originaleRésumé
Au cours des dernières années, l'étude des petites molécules (telles que les métabolites, les lipides et les médicaments) a suscité un intérêt croissant car celles-ci jouent un rôle important dans divers domaines de recherche, comme la biomédecine, les biotechnologies, les sciences de l'environnement ou encore la découverte de médicaments. La spectrométrie de masse (MS) est devenue une technologie de choix pour analyser ces composés de faible poids moléculaire. Cependant, l'analyse des petites molécules par désorption/ionisation laser assistée par matrice (MALDI, qui est une des techniques de référence en MS) peut s'avérer difficile en raison des interférences générées par la matrice dans la gamme des faibles m/z. C'est pourquoi de nouvelles techniques d'ionisation qui ne nécessitent pas de matrice organique ont été développées. Parmi ces alternatives, on retrouve la désorption/ionisation laser assistée par surface (SALDI), qui repose sur l’utilisation de surfaces nanostructurées pour promouvoir les processus de désorption/ionisation. Cependant, la mise en œuvre de cette nouvelle technique d'ionisation présente de nouveaux défis, notamment la compréhension des mécanismes fondamentaux qui régissent les processus SALDI et l'optimisation des facteurs expérimentaux. En particulier, la conception et l'optimisation des nanosubstrats ne sont pas simples, en raison de la large gamme de nanomatériaux disponibles (en termes de nature chimique et de morphologie), caractérisés par différentes propriétés, qui ont un impact sur la performance des expériences de SALDI MS. Ainsi, il est essentiel de trouver un modèle pour comparer les capacités des nanosubstrats SALDI, afin d'optimiser leurs caractéristiques sur base d'une référence commune. Dans ce contexte, les ions thermomètres ont été utilisés comme composés de référence pour tester de nouveaux instruments, de nouvelles méthodologies ou pour ajuster les paramètres instrumentaux. Dans cet article, nous passons en revue l'étude des ions thermomètres en SALDI MS, qui ont permis de rationaliser l'effet des facteurs expérimentaux (notamment la morphologie et la nature chimique du nanosubstrat, et les paramètres instrumentaux) sur la performance des analyses en SALDI MS. Ces études permettent la conception rationnelle de nanosubstrats optimisés pour des applications avancées, telles que l'imagerie SALDI MS, qui sera discutée à la fin de cet article.
Abstract
In recent years, there has been a growing interest in the study of small molecules (e.g. metabolites, lipids, drugs) as they play important roles in various research areas, including biomedicine, biotechnology, environmental science and drug discovery. Mass spectrometry (MS) has become a predominant technology to analyze these low molecular weight compounds. However, the analysis of small molecules by the established matrix-assisted laser desorption/ionization (MALDI) MS technique can be difficult because of the interference generated by the matrix in the low m/z range. Therefore, new innovating ionization techniques that do not require an organic matrix have been developed. Among these alternatives is surface-assisted laser desorption/ionization (SALDI) MS, which relies on nanostructured surfaces to promote the desorption/ionization process. Yet, the implementation of this novel ionization technique introduces new challenges, which include the understanding of the fundamental mechanisms that govern the SALDI process, and the optimization of the experimental factors. In particular, the design and optimization of the assisting nanosubstrates are not straightforward, because of the wide range of available nanomaterials (in terms of chemical nature and morphology), characterized by different properties, which impact the performance of the SALDI MS experiments. Thus, it is essential to find a model to compare the capabilities of the SALDI nanosubstrates, in order to optimize their characteristics based on the same reference. In this context, thermometer ions have been used as reference compounds to test novel instrumentation, new methodologies, or to tune instruments. In this article, we review the study of thermometer ions in SALDI MS, which have enabled to rationalize the effect of the experimental factors (i.e. the nanosubstrate morphology and chemical nature, and the instrumental settings) on the performance of SALDI MS experiments. These studies allow the rational design of optimized nanosubstrates for advanced applications, such as SALDI MS imaging, which will be discussed at the end of this article.
Tabla de contenidos
Manuscript received on May 29, 2022 and accepted on October 2, 2022
This article is distributed under the terms and conditions of the Creative Commons license CC-BY 4.0
1. Introduction
1Mass spectrometry (MS) is an analytical technique, which allows measuring the mass-to-charge ratios (m/z) of ions in the gas phase. Therefore, as neutral molecules will not be detected, the first step in mass spectrometry is to create charged particles from the sample molecules, and to transfer the ions into the gas phase. These steps take place in the ion source of the mass spectrometer. There are many different ionization techniques [1]. For example, ions can be formed by electron bombardment (i.e. electron ionization, EI) [2], by chemical reactions with an ionized reagent gas (i.e. chemical ionization, CI) [3], by evaporation of charged droplets (i.e. electrospray ionization, ESI) [4,5], or by irradiation with a laser beam (i.e. laser desorption/ionization (LDI) techniques), which may be supported by an assisting material, as in matrix-assisted laser desorption/ ionization (MALDI) [6-8], for example. The ions are then sent to a mass analyzer, which separates the ions according to their m/z. There are several properties (which are dependent on the m/z) that can be used to separate ions [1], including their time of flight in a drift tube (i.e. time-of-flight (TOF) analyzers), their cyclotron resonance frequency in a fixed magnetic field (i.e. Fourier-transform ion cyclotron resonance (FT-ICR) analyzers), or the stability of their trajectories in oscillating electric and/or magnetic fields (i.e. quadrupole, ion trap and orbitrap analyzers). The “sorted” ions finally reach the detector of the instrument, and the signal is converted into a mass spectrum showing the signal intensity of the ions as a function of their m/z value (Figure 1).
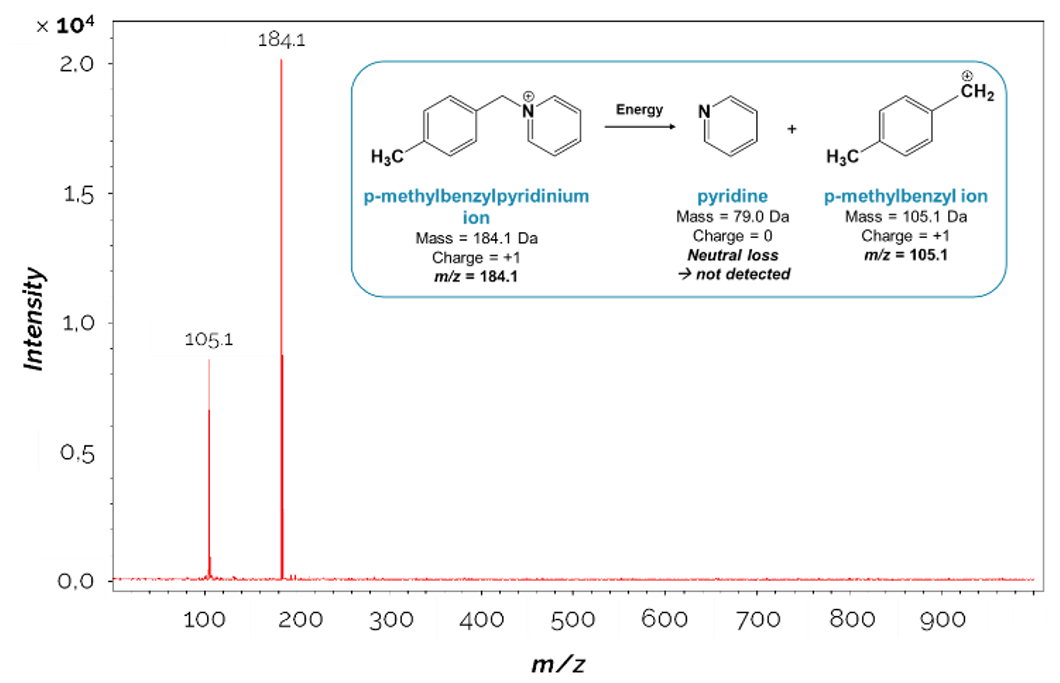
Figure 1.
Example of a mass spectrum acquired during the analysis of the p-methylbenzylpyridinium thermometer ion. The detected ions correspond to the p-methylbenzylpyridinium ion (m/z 184.1) and its fragment ion (m/z 105.1).
2Therefore, mass spectrometry can be used to identify compounds (by associating the m/z value to an exact mass, and then to a chemical formula), to study their structure and properties, and under certain conditions to quantify them. Thus, you will easily understand why mass spectrometry has become an essential technique, not only to answer fundamental questions, but also to help solving the challenges faced in biology [9-11], food safety [12], biomedical sciences [13-15], environmental sciences [16] and forensics [17], for example.
1.1. MALDI mass spectrometry
3Among all the ionization sources, soft ionization techniques, in particular ESI and MALDI, really brought about major change in the whole mass spectrometry area, by allowing the analysis of intact large biological molecules (such as proteins). The discovery of ESI by John B. Fenn and MALDI by Koichi Tanaka led to the 2002 Nobel Prize in Chemistry, highlighting the importance of these two MS techniques for the entire scientific community.
4The main difference between MALDI and previous ionization techniques based on laser irradiation is the use of a “matrix”, which is typically a low molecular weight organic compound, able to co-crystallize with the molecules present in the sample (Figure 2). The matrix plays a key role in MALDI MS by (i) protecting the analytes from direct laser irradiation (which limits their fragmentation), (ii) assisting the desorption (through the absorption of the laser energy and its transfer to the analytes), and (iii) promoting the ionization of the analytes (usually by (de)protonation).
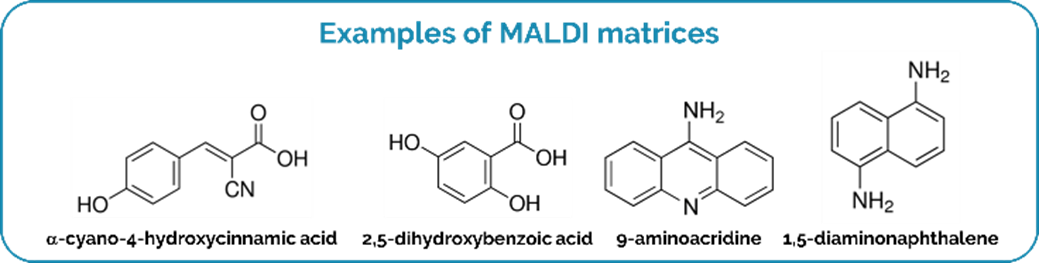
Figure 2.
Examples of MALDI matrices.
5Several theoretical models have been proposed to explain the desorption and ionization processes in MALDI, but the overall process remains unclear. Without going into details, two mechanisms have been proved to occur in parallel [18], namely (i) the “gas-phase protonation model”, which assumes that laser irradiation initially leads to the ablation of a cluster of neutral molecules, which are then charged by collisions with matrix ions in the gas phase, and (ii) the “Lucky Survivor model”, stating that all ions are preformed (i.e. they already exist in the sample before laser irradiation).
6However, while MALDI MS has revolutionized the study of macromolecules, this technique is not the most suitable for the analysis of small molecules (< 700 Da) [19-20]. Indeed, upon laser irradiation, the organic matrix is also desorbed, ionized, and potentially fragments and/or forms clusters, along with the desorption/ionization of the compounds of interest [21]. The desorption and ionization of all these species may lead to messy mass spectra, filled up with lots of interfering signals (especially in the low m/z range), which might hinder the detection of low molecular weight analytes. In other words, looking for the signals of the analytes of interest would be like looking for a needle in a haystack.
7Nevertheless, the study of small molecules (e.g. metabolites, lipids, drugs) has become a crucial task, mainly in life sciences. Indeed, small molecules have been found to play critical roles in biochemical processes, such as the development of a disease or intercellular communications. The study of small molecules in cells and biological tissues may help disease diagnosis and biomarker discovery, for example. Hence, alternative techniques that do not require the addition of an organic matrix have been developed to enable the analysis of small molecules.
1.2. SALDI mass spectrometry
8Among these alternatives is a technique called “SALDI” for surface-assisted laser desorption/ionization [21-22]. The acronym “SALDI” will be used throughout this article to describe all derived methodologies, including for example desorption/ionization on silicon (DIOS), nanostructure-initiator MS (NIMS), and nanopost array (NAPA) MS, so as not to confuse the readers with the intricate nomenclature associated with this group of techniques [21]. Instead of using organic matrices, SALDI MS relies on the use of nanostructured surfaces to assist the desorption/ionization of the analytes [21], [23-26]. The nanostructured surfaces used in SALDI MS can be classified into three general types: coatings of suspended nanomaterials (e.g. spherical nanoparticles, nanowires) [27,28], sputtered metal nanoclusters [29], and nanostructured solid substrates (e.g. porous silicon, nanopillar arrays) [30,31] (Figure 3).
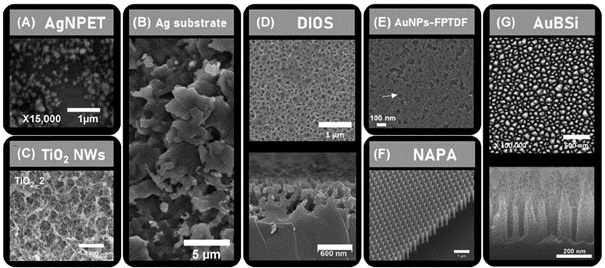
Figure 3.
SEM images of SALDI nanosubstrates. (A) silver nanoparticle‐enhanced target (AgNPET), (B) etched Ag substrate, (C) TiO2 nanowire (NWs) substrate, (D) DIOS nanosubstrate, (E) functionalized porous TiO2 film immobilized with gold nanoparticles (AuNPs‐FPTDF), (F) NAPA platform, and (G) gold‐coated black silicon substrate (AuBSi). Reprinted with permission from W.H. Müller et al. (2022) Mass Spectrom. Rev., 41(3), 373–420. Reference [21].
9One of the main advantages of the SALDI nanosubstrates lies in the near absence of interference generated in the low m/z region during their laser irradiation, which makes SALDI MS particularly interesting for the study of small molecules [21,32,33]. Moreover, the nanosubstrates have many other advantages that drive the development of the SALDI MS technique. These include for example their dual-polarity capabilities, which allow the analysis of the samples in both ionization modes with the same nanosubstrate (i.e. the nanosubstrate assists the formation of both positively and negatively charged species, which can be detected in the positive and negative ionization modes, respectively) [34,35]. This asset is particularly important for the study of molecule families that are detected in the two ionization modes, some preferentially in the positive and others in the negative ionization mode. This is for example the case of lipids [36]. The nanosubstrates also usually offer higher signal reproducibility compared to MALDI matrices [37,38]. Another major advantage is the controlled and adjustable design, especially of the solid nanostructured substrates, which enable to optimize the performance of the SALDI MS experiments. For example, the modification of the nanosubstrate surface chemistry allows improving the selectivity (e.g. the addition of a silver coating enhances selectivity towards long‐chain unsaturated hydrocarbons [39]), and the limits of detection (e.g. functionalization with superhydrophobic ligands leads to droplet confinement and enrichment of the analytes in a small spot [40,41]).
10Due to these many advantages, SALDI MS is developing rapidly, and has for example recently been used to visualize phospholipids in cancerous tissues in the perspective of cancer diagnosis [42], to analyze poisons in human serum [43], and to detect drugs in environmental samples [44]. However, the implementation of this novel ionization technique introduces new challenges, which include the understanding of the fundamental mechanisms that govern the SALDI process, and the optimization of the experimental factors.
2. How to optimize SALDI MS experiments?
11Before it can be applied in a practical context, for example, in a hospital or in a quality control laboratory, an analytical technique has to be mastered and optimized. However, the optimization of a mass spectrometry technique is not as straightforward as it seems. Indeed, the Holy Grail would be a MS technique that efficiently produces ions (i.e. with intense signal intensity and controlled fragmentation), which would be the ions of interest (i.e. selective method), detected with a good mass resolution and accuracy (allowing the confident analyte identification), and which would be detected even in very small quantities (i.e. good limits of detection), all with a good spatial resolution in the case of imaging experiments (see section 3). Of course, the optimization of all these parameters at once would be a utopia, and analysts usually have to decide on which analytical feature to base the optimization of the technique. In other words, the optimization consists in studying how changes in experimental factors affect a particular analytical parameter, such as the intensity or the reproducibility of the signal. The optimal conditions for the analysis can then be found by understanding the impact of the studied experimental factors.
12In SALDI MS, the probed experimental factors will include the instrumental settings but also the characteristics of nanosubstrates (e.g. bulk composition, surface chemistry and morphology), which play a key role in the SALDI process by absorbing the laser energy, enabling a rapid and sharp increase in the surface temperature. Yet, the design and optimization of the assisting nanosubstrates is not straightforward, because of the wide range of available nanomaterials (in terms of chemical nature and morphology), characterized by different properties, which impact the performance of the SALDI MS experiments. Thus, it is essential to find a model to compare the capabilities of the SALDI nanosubstrates, in order to optimize their characteristics based on the same reference. Moreover, the systematic study of the experimental factors affecting the performance of SALDI MS would be crucial not only for the rational selection and optimization of the SALDI nanostructured surfaces for analytical applications, but also from a fundamental point of view. Indeed, as for MALDI, the understanding of the fundamental mechanisms underlying the desorption and ionization processes in SALDI is still a significant challenge, although several studies have attempted to rationalize the SALDI mechanisms [21,45,46]. Briefly, the desorption is thought to mainly occur via thermal processes, including the rapid and highly localized heating of the nanosubstrate surface, coupled with heat confinement effects, resulting from the interaction of the laser with the substrate nanostructure [21,45,47]. Some other non-thermal processes may also help the analyte desorption, such as laser-induced surface restructuring or destruction [21,47]. On the other hand, ionization is thought to occur mainly via non-thermal processes, but remains misunderstood as it can be promoted by several phenomena, including charge transfers, photo-ionization reactions or surface melting/destruction [21,48].
2.1. Study of thermometer ions, the model analytes used as a reference
13Among all analytical parameters that can be investigated during MS experiments, the energy transfers are one of the most fundamental characteristics of an ion source [49]. These energy transfers occur between the “primary” energy source (e.g. the laser in SALDI MS) and the analytes. The energy can be transferred as kinetic (allowing efficient analyte desorption) and internal energy (which may induce fragmentation of the ions prior to detection [50]) [51]. The amount of internal energy that is transferred will drive the proportion of surviving “intact” molecular ions as opposed to their fragment ions formed in the ion source. Understanding these energy transfers between the laser, the nanosubstrate (as an intermediary) and the analytes is important to optimize the desorption efficiency (and thus the analyte signal intensity) but also to control the extent of fragmentation, which may be required (for structural characterization) or not (for molecular mass determination).
14In the 90’s, our laboratory, and in particular Edwin De Pauw and coworkers, developed a method for estimating the internal energy distribution of ions produced in soft ionization mass spectrometry experiments [52-54]. This method relies on the use of so-called “thermometer ions” (so named because their internal energy distribution can be characterized by means of a parameter having the dimension of a temperature) [55]. In practice, De Pauw and coworkers proposed to study the fragmentation of benzylpyridinium (BP) ions (Figure 4), used as thermometer ions, with the survival yield method [52-54].
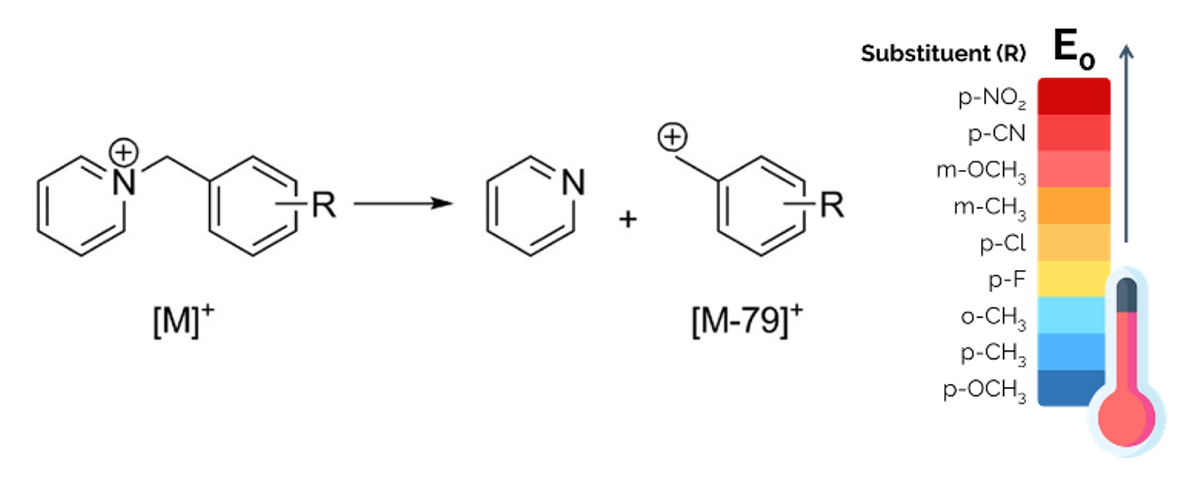
Figure 4.
(left) Fragmentation of the substituted benzylpyridinium “parent ion” leading to the formation of the “fragment ion” and neutral pyridine. (right) Scale of the dissociation energy threshold (E0) of the thermometer ions, depending on the substituent.
15Several compounds have been used as “thermometers ions”, such as transition-metal carbonyl complexes [56], tetraethylsilane ions [56,57], tetraphenylborate ions [58], and peptides [59], but the benzylpyridinium salts have several advantages over these other thermometers ions. First, benzylpyridinium ions are believed to have a very simple fragmentation pattern, which is an important requirement for thermometer ions [60]. Their fragmentation consists in the simple cleavage of the C-N bond between the benzyl C and the pyridine N, leading to the loss of neutral pyridine and the formation of the substituted benzyl cation as the only fragment [45], once the internal energy of the thermometer exceeds the dissociation energy threshold, E0 (Figure 4). This E0 threshold depends on the nature and position of the R substituent [61] (Figure 4). In other words, the observation of fragment ions in the mass spectra implies that enough internal energy has been transferred to the parent ions to cross the E0 energy threshold [60]. Second, benzylpyridinium salts are pre-ionized compounds, already existing as charged species associated with a counterion in the solid phase or as adsorbates. Their study can therefore focus on the desorption and the fragmentation processes, separately from the ionization process [62].
16Thermometer ions can thus be used to probe both the desorption efficiency and the internal energy transfer. The desorption efficiency (DE) is simply the sum of the signal intensities of the “parent” (e.g. [M]+ in Figure 4) and “fragment” (e.g. [M-79]+ in Figure 4) thermometer ions:
17DE = IParent + IFragment
18where IParent and IFragment indicate the intensity of the parent and fragment thermometer ions, respectively.
19On the other hand, the extent of internal energy transfer can be evaluated though the fragmentation of the “thermometer ion”. To this end, the relative proportion of the surviving intact “parent ions” of the thermometer to the total intensity of desorbed thermometer ions (both the parent and the fragment ions) is calculated. This proportion represents the survival yield (SY) of the thermometer ion, defined as:
20SY = IParent / (IParent + IFragment)
21Low survival yield values mean that the thermometer ion fragments a lot, and so that a high amount of internal energy has been transferred to the thermometer ion.
22Benzylpyridinium ions have been widely used as reference compounds in different MS techniques, to test novel instrumentation, new methodologies, or to tune the instruments [21,55,60,63]. Thus, BP ions are interesting model analytes for the study of the SALDI processes, and for the investigation of the experimental factors (specific to the nanosubstrate and to the instrumental settings) impacting the SALDI MS performance, in the perspective of optimizing both the characteristics of the nanosubstrates and the experimental conditions, based on a common reference. In particular, the investigation of the desorption and fragmentation of the BP ions during SALDI MS experiments allowed to understand (at least partially) how the nanosubstrate morphology and chemical nature affect the SALDI MS analyses, and to monitor the optimal instrumental settings for efficient desorption and controlled fragmentation of the analytes.
2.2. Investigation of the nanosubstrate morphology
23As previously said, the nanosubstrate plays a significant role in SALDI MS analyses, thus the optimization of its characteristics is essential. For instance, the nanosubstrate morphology has been reported to be a significant factor affecting the desorption efficiency and the degree of fragmentation in SALDI MS [58,64-66]. Even if optimal SALDI performance is often achieved by a combination of factors acting synergistically rather than a particular characteristic of the nanosubstrate [67], some structural aspects proved to be particularly crucial. The study of benzylpyridinium ions has helped to understand the influence of the nanosubstrate structural properties on the SALDI processes. The key aspects, including the dimensions [68,69] and dimensionality [69-72] of the nanostructures, their orientation (e.g. nanowires in a random cross-linked network vs. aligned nanowires) [68,70], and the surface voidage [66] (i.e. the percentage of air on the surface of the nanostructure [58]), are mainly responsible for efficient thermal energy confinement, which is required to ensure a drastic increase in the surface temperature, promoting efficient analyte desorption [21,22,47]. For example, in one-dimensional structures such as nanorods and nanotubes, the heat conduction is hampered as the thermal energy can only dissipate in the axial direction, which leads to energy confinement and thus, high desorption efficiency [70]. On the contrary, in two or three-dimensional nanostructures such as nanosheets, the thermal energy is not confined, and can freely dissipate in multiple directions, leading to a low desorption efficiency [70]. The increase in the surface temperature can also be caused by a reduction of the surface thermal conductivity by increasing the surface voidage [58,67], for example by increasing the porosity of the substrate [58,67], or by increasing the wall-to-wall distance in nanowall substrates [66]. Finally, the dimensionality also impacts the energy transfers in the plume (i.e. all material vaporized from the sample, including analyte ions, sometimes nanosubstrate ions, and neutrals). In one-dimensional nanostructures (such as nanopores in porous silicon substrates), the plume expansion is confined in one direction in the pore structure, which maintains a relatively high plume pressure, and thus promotes energy transfers [70]. On the contrary, when the plume can expanse in three directions, the plume density drops rapidly and the energy transfers are limited [70, 73]. As an example, the survival yields of benzylpyridinium thermometer ions, acquired in SALDI MS with porous silicon were found to be lower than in MALDI MS with α-cyano-4-hydroxycinnamic acid (CHCA), and SALDI MS with a network of silicon nanowires (Figure 5) [72].
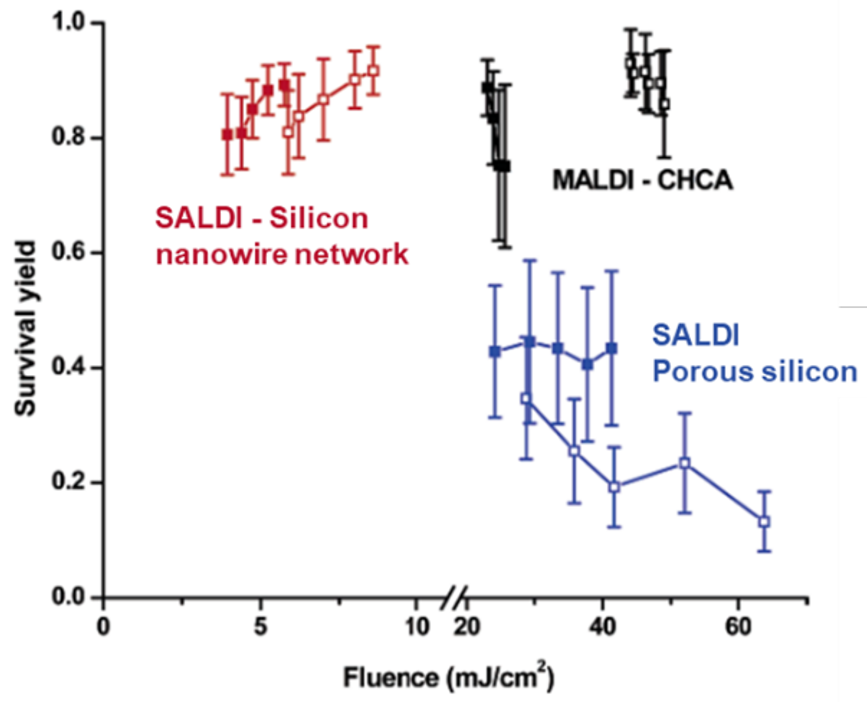
Figure 5.
Survival yield of BP thermometer ions as a function of the laser fluence, acquired using CHCA in MALDI MS, and a silicon nanowire network and porous silicon in SALDI MS. Adapted with permission from G. Luo et al. (2006) J. Phys. Chem. B, 110, 13381–13386 © 2006 American Chemical Society. Reference [70].
2.3. Investigation of the nanosubstrate chemical nature
24The chemical nature of the nanosubstrates (including the bulk composition and the surface chemistry) also has a significant impact on the SALDI performance, and can be easily tuned. For example, several functional groups can be used to modify the surface chemistry of the nanosubstrates by functionalization with specific ligands (e.g. silane ligands can react with silanol groups (Si-OH) at the surface of silicon substrates, while thiol ligands (S-H) can be used to derivatize gold surfaces). However, the modification of the chemical nature of the nanosubstrates implies changes in the substrate properties, which will ultimately impact the performance of the SALDI MS experiments [48,62,74-81]. In this context, the study of thermometer ions also allowed to rationalize the effects of the nanosubstrate chemical nature on the desorption efficiency and survival yield examined in SALDI MS.
25For instance, the surface chemistry of solid nanostructured substrates and nanoparticles is commonly modified with (super)hydrophobic coatings (e.g. functionalization with (per)fluorinated ligands) [41,82,83]. The superhydrophobic nature of the surface leads to droplet confinement, and subsequent enrichment of the analytes in a very small area on the nanosubstrate surface, which ultimately improve the limits of detection [73]. In addition, this functionalization with superhydrophobic ligands modifies other properties, which have notably been studied using thermometer ions. For example, the superhydrophobic ligands decrease the surface wettability, which leads to a less complete penetration of the analytes into the nanopores. The analytes thus need less energy to be “extracted”, which leads to higher desorption efficiency [48]. The functionalization with superhydrophobic ligands also decreases the interaction energy between the surface and the analytes, which also leads to higher desorption efficiency [48,58,73,74]. Moreover, the ligands also act as energy moderators, by absorbing a part of the laser energy transferred to the analytes, which leads to higher survival yields (i.e. less fragmentation), as shown in Figure 6 [58,62,73,74].
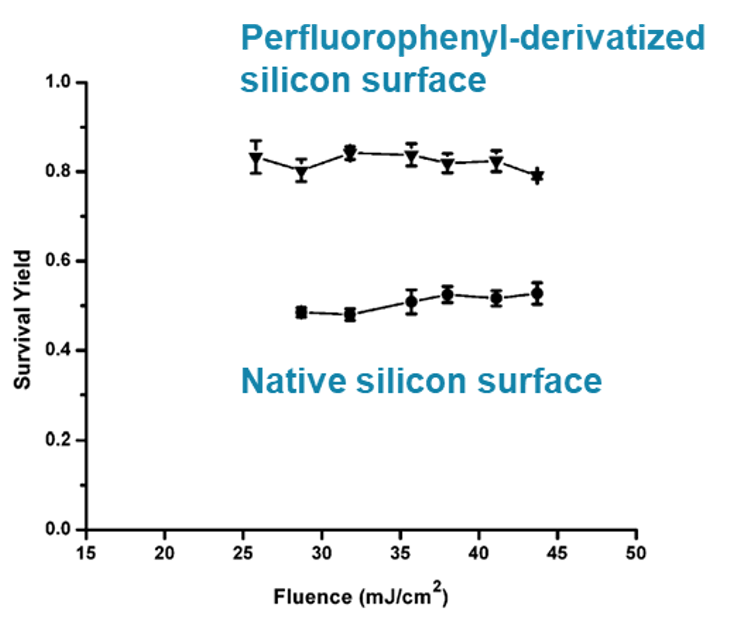
Figure 6.
Survival yield of BP thermometer ions as a function of the laser fluence, acquired using perfluorophenyl-derivatized silicon microcolumn arrays and native silicon microcolumn arrays. Adapted with permission from J.A. Stolee et al. (2010) J. Phys. Chem. C, 114(12), 5574–5581 © 2010 American Chemical Society. Reference [62].
26Changes in the chemical nature can also impact the UV/Visible absorbance of the nanosubstrate. It is known that the desorption is mainly a thermal process engendered by the heating of the substrate surface upon absorption of the laser energy (the common laser wavelength is 355 nm, in the UV region of the electromagnetic spectrum) [84]. A high absorbance in the UV/Visible is therefore required in SALDI MS. For example, the addition of gold on steel [85] and TiO2 [75] substrates was shown to lead to an increased desorption efficiency and energy transfer, in concordance with an increased UV/Visible absorbance of the substrates. The SY method also allowed to study the influence of the nature of noble metal nanoparticles on the internal energy transfers [78]. Again, increased energy transfer (i.e. lower survival yield) was correlated to higher UV/Visible absorbance of the nanoparticles (Figure 7) [78].
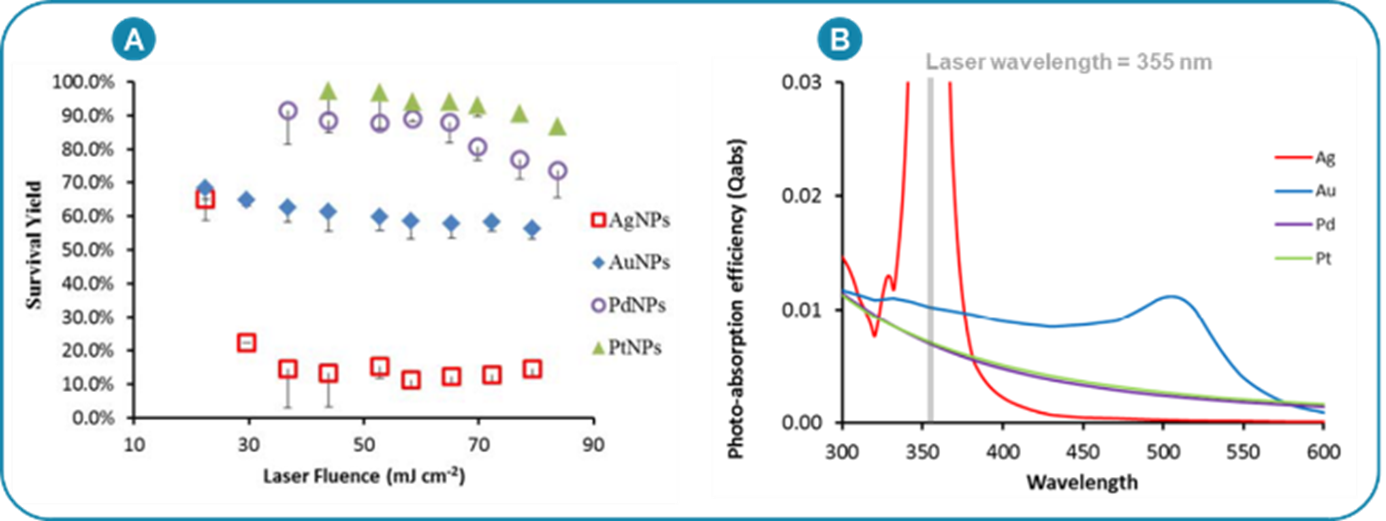
Figure 7.
(A) Survival yield of BP thermometer ions as a function of the laser fluence, acquired using different noble metal nanoparticles, composed of silver, gold, palladium and platinum, respectively. (B) Photo-absorption of the noble metal nanoparticles. Adapted with permission from K. Ng et al. (2015) J. Phys. Chem. C, 119, 23708–23720 © 2015 American Chemical Society. Reference [78].
2.4. Investigation of the instrumental settings
27Finally, even with model analytes and optimized nanosubstrates, the SALDI MS analysis can go wrong if the instrumental settings are not properly tuned. Indeed, the desorption efficiency and fragmentation are affected by the experimental conditions and the instrumental settings [86]. Some can be adjusted by the operator (e.g. laser power), while others are imposed by the instrumentation (e.g. geometry of the ion source). Recently, benzylpyridinium ions have been analyzed in our laboratory using silver-capped silicon nanopillar arrays as nanosubstrates, in the perspective of clarifying the impact of the instrumental settings in SALDI MS experiments. Some operational parameters of the RapifleX MALDI TOF-TOF mass spectrometer were examined, namely the laser power, the pixel size, the laser pulse repetition rate, the number of accumulated laser shots, and the duration of the pulsed delay extraction (which is a time delay between the ionization pulse and an accelerating voltage pulse used to temporally focus the ions produced in the ion source in order to improve the mass resolution). Among the studied parameters, almost all were found to have an impact both on the desorption efficiency and on the fragmentation of the BP ions, which underlines the importance of the instrumental settings optimization.
28For example, the laser power was found to have a considerable impact on the desorption efficiency (Figure 8A), which was expected as the laser power is the operating parameter that enables to modulate the primary energy deposited on the sample. The increasing amount of energy transferred from the laser to the nanosubstrate (and then, to the thermometer ions) when the laser power is increased explains the raising trend of the desorption efficiency (Figure 8A) [37,38]. Thus, during the optimization procedure, operators would be tempted to increase the laser power to get a high signal intensity. However, as shown in Figure 8A, beyond ~ 60-65% of the laser power, the behavior of the desorption efficiency changes. This change can be explained by the destruction of the irradiated nanosubstrate at high laser power values. This was confirmed by the dramatic increase in the intensity of silver ions (107Ag+) emitted by the silver-capped silicon nanopillars after laser irradiation at laser power values above ~ 60-65% (Figure 8B). Thus, the optimization of the laser power has to be a matter of trade-off between signal intensity, analyte fragmentation and substrate destruction. On the other hand, the study showed that the optimization of other instrumental settings (i.e. pixel size, pulsed delay extraction and number of laser shots) could be done by focusing only on the signal intensity of the ion of interest, as these settings have only a minimal effect on the fragmentation.
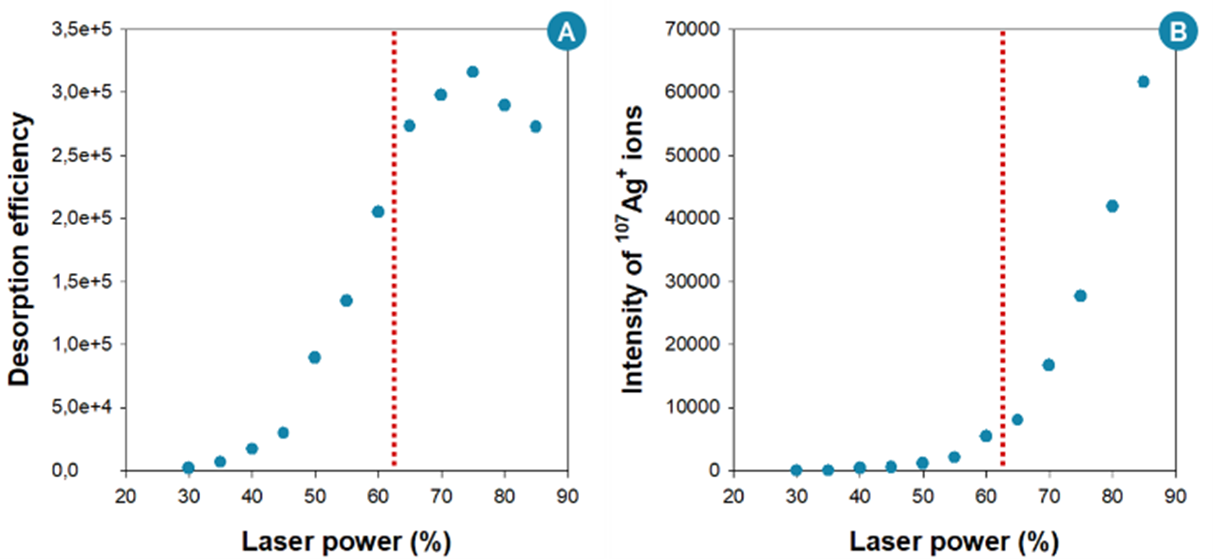
Figure 8.
(A) Desorption efficiency of BP thermometer ions as a function of the laser power, acquired using silver-capped silicon nanopillar arrays. (B) Intensity of 107Ag+ ions as a function of the laser power, produced in parallel with benzylpyridinium ions during the silver-capped silicon nanopillar substrate irradiation.
3. Towards SALDI mass spectrometry imaging
29The ultimate goal of the optimization of the SALDI nanosubstrates is their use in practical applications, in order to meet the modern challenges faced in various research areas, such as the biomedical field [29,87] and forensics [88,89]. The use of the SALDI nanosubstrates in mass spectrometry imaging (MSI) analyses [90-92] is also a promising application. The growing interest in MSI is linked to the new valuable information brought by the imaging aspect, additionally to the analyte detection and identification offered by conventional mass spectrometry. Indeed, mass spectrometry imaging experiments allow visualizing the spatial distribution of the molecules present in the samples [13,90,92], which can be as complex as biological tissue sections [13,93], fingerprints [94,95], bacterial colonies and cells [96].
30During MSI experiments, the spatial distributions of the compounds of interest are determined by acquiring a set of position-correlated mass spectra. Thousands of mass spectra are usually acquired over the entire analyzed area, and constitute the pixels that will be used to reconstruct the ion image of the sample. The ion image is the intensity map of a specific m/z signal, and represents the spatial distribution of the signal intensity associated with that specific m/z value (Figure 9).
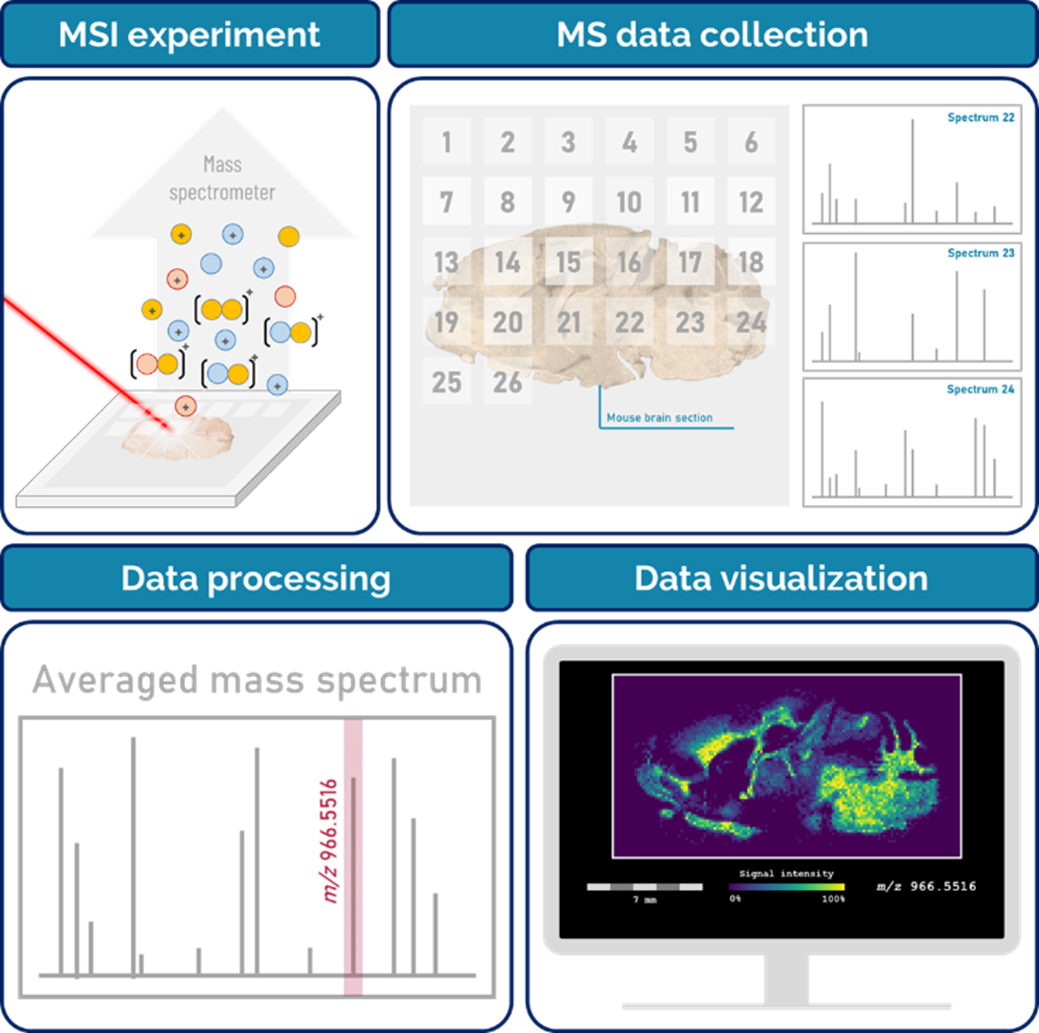
Figure 9.
Schematic principle of mass spectrometry imaging experiments.
31In particular, the emergent SALDI MSI, requiring no organic matrix, represents a powerful alternative to the reference MALDI MSI technique, especially for the imaging of small molecular species. Indeed, as previously said, MALDI MS suffers from matrix-related limitations, and some issues are specifically impacting imaging analyses. For example, if the matrix is not applied homogeneously on the sample, the uneven matrix-analyte co-crystallization may lead to the production of “hot spots” (i.e. sample areas with more intense signals) and thus, to a lack of reproducibility [97,98]. The “wet application” of the matrix (i.e. when the matrix solution is sprayed on the sample) may also lead to the delocalization of the analytes within the sample, and thus affect the accuracy of the ion images, which in turn can lead to misinterpretation of the MSI results [99-101]. On the contrary, the nanostructured substrates used in SALDI MSI generally produce more reproducible signal intensities [37,38], and some sample preparation procedures (e.g. metal sputtering) allow to perform SALDI MSI at high spatial resolution (with no analyte delocalization) [21,39,102].
32Therefore, SALDI mass spectrometry imaging opens up new opportunities to provide unequaled insights, highly valuable in various scientific disciplines. For example, the visualization of the spatial distributions of small molecules in biological tissue samples may facilitate clinical diagnosis by identifying the regions where the pathology is delimited (Figure 10). Interested readers are invited to consult the review written by Müller W.H. et al. [21] to discover other SALDI MSI applications.
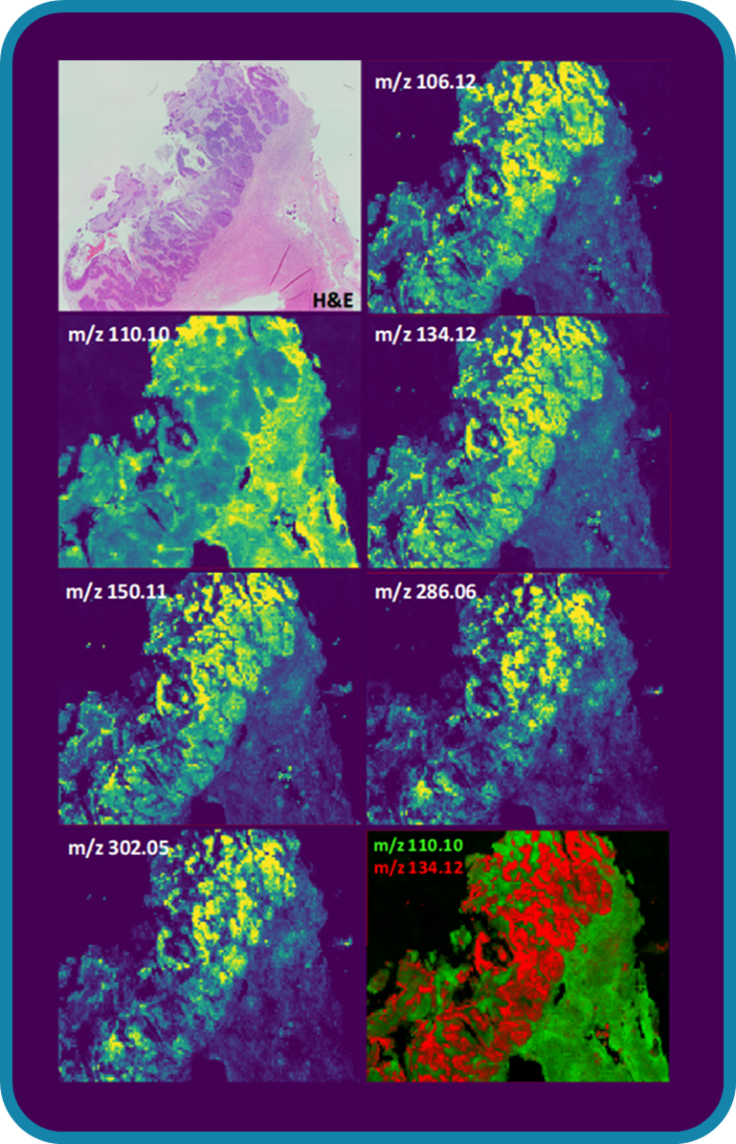
Figure 10.
H&E stain and SALDI mass spectrometry imaging of a cancerous ovarian tissue section highlighting the diseased areas. Credits: Alexandre Verdin
33However, the development of these advanced applications require to gain additional insights into the key mechanisms involved in the SALDI process. As seen throughout this article, thermometer ions can be valuable allies for the study of SALDI MS. Indeed, the studies of benzylpyridinium ions have enabled to understand general trends regarding the impact of the nanosubstrate characteristics and instrumental settings on the SALDI MS performance. However, benzylpyridinium ions are only model compounds, and are not fully representative of the analytes that will be studied in actual samples (which may not behave exactly like the thermometer ions). In any case, it will also be necessary to optimize the analysis of the sample of interest. Moreover, the studies of benzylpyridinium ions generally focused on the analysis of dried-droplets of the pure thermometer ion solution. Actual samples are usually much more complex, especially in the context of imaging analyses. The samples may contain hundreds or even thousands of analytes, characterized by very different properties (e.g. mass, polarity, ionization efficiency). These applications will therefore need their own rigorous optimization procedures. The quest for the Holy Grail is still ongoing.
4. Conclusion
34Over the past few years, surface-assisted laser desorption/ionization (SALDI) mass spectrometry (MS) has gained growing attention, mainly for the study of small molecules, as this technique does not suffer from matrix-related issues. The nanostructured substrates used in SALDI MS, which play a key role in the desorption/ionization processes, are responsible for the unique capabilities of this technique. Therefore, the optimization of the nanosubstrate characteristics (e.g. bulk composition, surface chemistry and morphology) is of prime importance. Yet, the optimization of the nanosubstrates may be challenging due to the diversity of available nanomaterials, characterized by a wide range of physico-chemical properties, which impact the SALDI MS performance. Therefore, a model to compare the capabilities of all SALDI nanosubstrates, in order to optimize their characteristics based on a common reference was highly desired. In this context, the study of the desorption and fragmentation of benzylpyridinium thermometer ions has provided some answers, by highlighting significant features (related to the nanosubstrate morphology and chemical nature, and to the instrumental settings) impacting the SALDI MS performance, and by rationalizing these impacts. These studies allow to optimize the engineering of the nanosubstrates and to find the appropriate experimental conditions for efficient desorption and controlled fragmentation in SALDI MS. The ultimate goal of the optimization of the SALDI nanosubstrates is their use in practical applications, such as SALDI MS imaging analyses, in order to meet the modern challenges. However, for these advanced applications, further optimization procedures are still required.
Acknowledgments
35W.H.M. acknowledges financial support from the F.R.S.‐FNRS. W.H.M. also thank her promoter Gauthier Eppe, her supervisors and colleagues Edwin De Pauw, Cedric Malherbe and Alexandre Verdin, and Laurane Gilliard for their valuable comments, which enabled the improvement of the manuscript.
Further information
36ORCID identifiers of author
37Wendy H. Müller 0000-0002-9216-6213
Bibliographie
[1] Y.-H. Tsai, M. R. F, D. M. Drexler, R. A. Yost, and T. J. Garrett, “Ionization sources and mass analyzers in MS imaging,” Bioanalysis, vol. 7, no. 20, pp. 2629–2637, 2015. doi: 10.4155/bio.15.187
[2] N. Mirsaleh-Kohan, W. D. Robertson, and R. N. Compton, “Electron ionization time‐of‐flight mass spectrometry: Historical review and current applications,” Mass Spectrom. Rev., vol. 27, pp. 237–285, 2008. doi: 10.1002/mas.20162
[3] A. G. Harrison, Chemical Ionization Mass Spectrometry, 2nd edition. New York: Routledge, 1992.
[4] J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong, and C. M. Whitehouse, “Electrospray Ionization for Mass Spectrometry of Large Biomolecules,” Science, vol. 246, p. 4926, 1989. doi: 10.1126/science.2675315
[5] S. Banerjee and S. Mazumdar, “Electrospray Ionization Mass Spectrometry: A Technique to Access the Information beyond the Molecular Weight of the Analyte,” Int. J. Anal. Chem., vol. 2012, 282574, 2012. doi: 10.1155/2012/282574
[6] R. Zenobi and R. Knochenmuss, “Ion formation in MALDI mass spectrometry,” Mass Spectrom. Rev., vol. 17, pp. 337–366, 1998. doi: 10.1002/(SICI)1098-2787(1998)17:5<337::AID-MAS2>3.0.CO;2-S
[7] K. Dreisewerd, “The Desorption Process in MALDI,” Chem. Rev., vol. 103, pp. 395–425, 2003. doi: 10.1021/cr010375i
[8] R. Knochenmuss and R. Zenobi, “MALDI Ionization: The Role of In-Plume Processes,” Chem. Rev., vol. 103, pp. 441–452, 2003. doi: 10.1021/cr0103773
[9] B. A. Boughton, D. Thinagaran, D. Sarabia, A. Bacic, and U. Roessner, “Mass spectrometry imaging for plant biology: a review,” Phytochem. Rev., vol. 15, pp. 445–488, 2016. doi: 10.1007/s11101-015-9440-2
[10] P. Lössl, M. Van De Waterbeemd, and A. J. R. Heck, “The diverse and expanding role of mass spectrometry in structural and molecular biology,” EMBO J., vol. 35, no. 24, pp. 2634–2657, 2016. doi: 10.15252/embj.201694818
[11] M. M. Gessel, J. L. Norris, and R. M. Caprioli, “MALDI imaging mass spectrometry: Spatial molecular analysis to enable a new age of discovery,” J. Proteomics, vol. 107, pp. 71–82, 2014. doi: 10.1016/j.jprot.2014.03.021
[12] I. Dominguez, A. Frenich, and R. Romero-Gonzalez, “Mass spectrometry approaches to ensure food safety,” Anal. Methods, vol. 12, no. 9, pp. 1148–1162, 2020. doi: 10.1039/C9AY02681A
[13] K. Chughtai and R. M. A. Heeren, “Mass Spectrometric Imaging for Biomedical Tissue Analysis,” Chem. Rev., vol. 110, no. 5, pp. 3237–3277, 2010. doi: 10.1021/cr100012c
[14] K. Schwamborn, M. Kriegsmann, and W. Weichert, “MALDI imaging mass spectrometry — From bench to bedside,” Biochim. Biophys. Acta, vol. 1865, pp. 776–783, 2017. doi: 10.1016/j.bbapap.2016.10.014
[15] A. G. Woods and C. C. Darie, Advancements of Mass Spectrometry in Biomedical Research, 1st ed. Springer, 2014.
[16] A. T. Lebedev, “Environmental Mass Spectrometry,” Annu. Rev. Anal. Chem., vol. 6, pp. 163–189, 2013. doi: 10.1146/annurev-anchem-062012-092604
[17] H. M. Brown, T. J. McDaniel, P. W. Fedick, and C. C. Mulligan, “The current role of mass spectrometry in forensics and future prospects,” Anal. Methods, vol. 12, no. 32, pp. 3974–3997, 2020. doi: 10.1039/D0AY01113D
[18] T. W. Jaskolla and M. Karas, “Compelling Evidence for Lucky Survivor and Gas Phase Protonation: The Unified MALDI Analyte Protonation Mechanism,” J. Am. Soc. Mass Spectrom., vol. 22, pp. 976–988, 2011. doi: 10.1007/s13361-011-0093-0
[19] C. D. Calvano, A. Monopoli, T. R. I. Cataldi, and F. Palmisano, “MALDI matrices for low molecular weight compounds: an endless story?,” Anal. Bioanal. Chem., vol. 410, pp. 4015–4038, 2018. doi: 10.1007/s00216-018-1014-x
[20] N. Bergman, D. Shevchenko, and J. Bergquist, “Approaches for the analysis of low molecular weight compounds with laser desorption/ionization techniques and mass spectrometry,” Anal. Bioanal. Chem., vol. 406, pp. 49–61, 2014. doi: 10.1007/s00216-013-7471-3
[21] W. H. Müller, A. Verdin, E. De Pauw, C. Malherbe, and G. Eppe, “Surface-assisted laser desorption/ionization mass spectrometry imaging: A review,” Mass Spectrom. Rev., vol. 41, no. 3, pp. 373–420, 2022. doi: 10.1002/mas.21670
[22] K. P. Law and J. R. Larkin, “Recent advances in SALDI-MS techniques and their chemical and bioanalytical applications,” Anal. Bioanal. Chem., vol. 399, no. 8, pp. 2597–2622, 2011. doi: 10.1007/s00216-010-4063-3
[23] H. He, Z. Guo, Y. Wen, S. Xu, and Z. Liu, “Recent advances in nanostructure/nanomaterial-assisted laser desorption/ionization mass spectrometry of low molecular mass compounds,” Anal. Chim. Acta, vol. 1090, pp. 1–22, 2019. doi: 10.1016/j.aca.2019.08.048
[24] M. Lu, X. Yang, Y. Yang, P. Qin, X. Wu, and Z. Cai, “Nanomaterials as Assisted Matrix of Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for the Analysis of Small Molecules,” Nanomaterials, vol. 7, no. 87, 2017. doi: 10.3390/nano7040087
[25] A. M. Dattelbaum and S. Iyer, “Surface-assisted laser desorption/ionization mass spectrometry,” Expert Rev. Proteomics, vol. 3, no. 1, pp. 153–161, 2006. doi: 10.1586/14789450.3.1.153
[26] P. A. Kuzema, “Small Molecule Analysis by Surface Assisted Laser Desorption/Ionization Mass Spectrometry,” J. Anal. Chem., vol. 66, no. 13, pp. 1227–1242, 2011. doi: 10.1134/S1061934811130065
[27] H. N. Abdelhamid, “Nanoparticle-based surface assisted laser desorption ionization mass spectrometry: a review,” Microchim. Acta, vol. 186, no. 682, 2019. doi: 10.1007/s00604-019-3770-5
[28] R. Pilolli, F. Palmisano, and N. Cioffi, “Gold nanomaterials as a new tool for bioanalytical applications of laser desorption ionization mass spectrometry,” Anal. Bioanal. Chem., vol. 402, no. 2, pp. 601–623, 2012. doi: 10.1007/s00216-011-5120-2
[29] M. Dufresne, N. H. Patterson, N. Lauzon, and P. Chaurand, “Assessing the Potential of Metal-Assisted Imaging Mass Spectrometry in Cancer Research,” Adv. Cancer Res., vol. 134, pp. 67-84, 2017. doi: 10.1016/bs.acr.2016.11.003
[30] Y. E. Silina and D. A. Volmer, “Nanostructured solid substrates for efficient laser desorption/ionization mass spectrometry (LDI-MS) of low molecular weight compounds,” Analyst, vol. 138, pp. 7053–7065, 2013. doi: 10.1039/C3AN01120H
[31] S. A. Iakab, P. Rafols, M. García-altares, O. Yanes, and X. Correig, “Silicon-Based Laser Desorption Ionization Mass Spectrometry for the Analysis of Biomolecules : A Progress Report,” Adv. Funct. Mater., vol. 29, p. 1903609, 2019. doi: 10.1002/adfm.201903609
[32] Z. Lin and Z. Cai, “Negative ion laser desorption/ionization time-of-flight mass spectrometric analysis of small molecules by using nanostructured substrate as matrices,” Mass Spectrom. Rev., vol. 37, pp. 681–696, 2018. doi: 10.1002/mas.21558
[33] H. Huang, D. Ouyang, and Z. Lin, “Recent Advances in Surface-Assisted Laser Desorption/Ionization Mass Spectrometry and Its Imaging for Small Molecules,” J. Anal. Test., pp. 1–18, 2022. doi: 10.1007/s41664-022-00211-5
[34] X.-N. Wang et al., “Porous TiO2 Film Immobilized with Gold Nanoparticles for Dual- Polarity SALDI MS Detection and Imaging,” ACS Appl. Mater. Interfaces, vol. 12, no. 38, pp. 42567–42575, 2020. doi: 10.1021/acsami.0c12949
[35] W. H. Müller et al., “Dual polarity SALDI FT-ICR MS imaging and Kendrick Mass Defect data filtering for lipid analysis,” Anal. Bioanal. Chem., vol. 413, no. 10, pp. 2821–2830, 2021. doi: 10.1007/s00216-020-03020-w
[36] W. H. Müller, E. De Pauw, J. Far, C. Malherbe, and G. Eppe, “Imaging lipids in biological samples with surface-assisted laser desorption/ionization mass spectrometry: A concise review of the last decade,” Prog. Lipid Res., vol. 83, no. July, p. 101114, 2021. doi: 10.1016/j.plipres.2021.101114
[37] M. Hasan et al., “Desorption ionization using through-hole alumina membrane offers higher reproducibility than DHB, a widely used matrix in FT-ICR mass spectrometry imaging analysis,” Rapid Commun. Mass Spectrom., p. e9076, 2021. doi: 10.1002/rcm.9076
[38] L. Krasny, M. Strnadova, K. Lemr, and V. Havlicek, “Lateral resolution in NALDI MSI: back to the future,” Anal. Bioanal. Chem., vol. 407, pp. 2141–2147, 2015. doi: 10.1007/s00216-014-8294-6
[39] M. Dufresne, A. Thomas, J. Breault-Turcot, J.-F. J.-F. Masson, and P. Chaurand, “Silver-Assisted Laser Desorption Ionization For High Spatial Resolution Imaging Mass Spectrometry of Olefins from Thin Tissue Sections,” Anal. Chem., vol. 85, pp. 3318–3324, 2013. doi: 10.1021/ac3037415
[40] M. Dupre, C. Enjalbal, S. Cantel, J. Martinez, and R. Boukherroub, “Investigation of Silicon-Based Nanostructure Morphology and Chemical Termination on Laser Desorption Ionization Mass Spectrometry Performance,” Anal. Chem., vol. 84, pp. 10637–10644, 2012. doi: 10.1021/ac3021104
[41] Q. Zhu, F. Teng, Z. Wang, Y. Wang, and N. Lu, “Confining analyte droplets on visible Si pillars for improving reproducibility and sensitivity of SALDI-TOF MS,” Anal. Bioanal. Chem., vol. 411, pp. 1135–1142, 2019. doi: 10.1007/s00216-018-01565-5
[42] X. Liu et al., “Tissue Imprinting on 2D Nano flakes-Capped Silicon Nanowires for Lipidomic Mass Spectrometry Imaging and Cancer Diagnosis,” ACS Nano, vol. 16, pp. 6916–6928, 2022. doi: 10.1021/acsnano.2c02616
[43] S. A. Al-Sayed, M. O. Amin, and E. Al-Hetlani, “SALDI Substrate-Based FeNi Magnetic Alloy Nanoparticles for Forensic Analysis of Poisons in Human Serum,” Molecules, vol. 27, p. 2720, 2022. doi: 10.3390/molecules27092720
[44] S. Dou, J. Lu, Q. Chen, C. Chen, and N. Lu, “High-density Si nanopillars modified with Ag nanoislands: Sensitive SALDI-MS chip for sulfonamides,” Sensors Actuators B. Chem., vol. 364, p. 131846, 2022. doi: 10.1016/j.snb.2022.131846
[45] R. A. Picca, C. D. Calvano, N. Cioffi, and F. Palmisano, “Mechanisms of Nanophase-Induced Desorption in LDI-MS. A Short Review,” Nanomaterials, vol. 7, no. 75, 2017. doi: 10.3390/nano7040075
[46] J. A. Stolee, B. N. Walker, V. Zorba, R. E. Russo, and A. Vertes, “Laser – nanostructure interactions for ion production,” Phys. Chem. Chem. Phys., vol. 14, pp. 8453–8471, 2012. doi: 10.1039/C2CP00038E
[47] K. Song and Q. Cheng, “Desorption and ionization mechanisms and signal enhancement in surface assisted laser desorption ionization mass spectrometry (SALDI-MS),” Appl. Spectrosc. Rev., vol. 55, no. 3, pp. 220–242, 2020. doi: 10.1080/05704928.2019.1570519
[48] G. Luo, Y. Chen, G. Siuzdak, and A. Vertes, “Surface Modification and Laser Pulse Length Effects on Internal Energy Transfer in DIOS,” J. Phys. Chem. B, vol. 109, no. 202, pp. 24450–24456, 2005. doi: 10.1021/jp054311d
[49] E. R. Stephens, M. Dumlao, D. Xiao, D. Zhang, and W. A. Donald, “Benzylammonium Thermometer Ions: Internal Energies of Ions Formed by Low Temperature Plasma and Atmospheric Pressure Chemical Ionization,” J. Am. Soc. Mass Spectrom., vol. 26, no. 12, pp. 2081–2084, 2015. doi: 10.1007/s13361-015-1272-1
[50] R. Rahrt, T. Auth, M. Demireva, P. B. Armentrout, and K. Koszinowski, “Benzhydrylpyridinium Ions : A New Class of Thermometer Ions for the Characterization,” Anal. Chem., vol. 91, pp. 11703–11711, 2019. doi: 10.1021/acs.analchem.9b02257
[51] V. Gabelica, E. Schulz, and M. Karas, “Internal energy build‐up in matrix‐assisted laser desorption ionization,” J. Mass Spectrom., vol. 39, pp. 579–593, 2004. doi: 10.1002/jms.651
[52] C. Collette and E. De Pauw, “Calibration of the Internal Energy Distribution of Ions Produced by Electrospray,” Rapid Commun. Mass Spectrom., vol. 12, pp. 165–170, 1998. doi: 10.1002/(SICI)1097-0231(19980227)12:4<165::AID-RCM140>3.0.CO;2-1
[53] E. De Pauw, G. Pelzer, J. Marien, and P. Natalis, “Internal Energy Distribution of Ions Emitted in Secondary Ion,” in Ion Formation from Organic Solids (IFOS III), Berlin Heidelberg: Springer, 1986, pp. 103–108.
[54] F. Derwa, E. De Pauw, and P. Natalis, “New Basis for a Method for the Estimation of Secondary Ion Internal Energy Distribution in ‘Soft’ Ionization Techniques,” Org. Mass Spectrom., vol. 26, pp. 117–118, 1991. doi: 10.1002/oms.1210260215
[55] J.-F. Greisch, V. Gabelica, F. Remacle, and E. De Pauw, “Thermometer ions for matrix-enhanced laser desorption/ionization internal energy calibration,” Rapid Commun. Mass Spectrom., vol. 17, pp. 1847–1854, 2003. doi: 10.1002/rcm.1124
[56] V. H. Wysocki, H. I. Kenttämaa, and R. G. Cooks, “Internal energy distributions of isolated ions after activation by various methods,” Int. J. Mass Spectrom. Ion Process., vol. 75, no. 2, pp. 181–208, 1987. doi:10.1016/0168-1176(87)83054-9
[57] R. D. Voyksner and T. Pack, “Investigation of collisional‐activation decomposition process and spectra in the transport regions of an electrospray single‐quadrupole mass spectrometer,” Rapid Commun. mass Spectrom., vol. 5, no. 6, pp. 263–268, 1991. doi: 10.1002/rcm.1290050604
[58] Q. Zhu et al., “Investigation of surface morphology on ion desorption in SALDI-MS on tailored silicon nanopillar arrays,” J. Phys. Chem. C, vol. 124, pp. 2450–2457, 2020. doi: 10.1021/acs.jpcc.9b09520
[59] J. Sztaray, A. Memboeuf, L. Drahos, and K. Vékey, “Leucine Enkephalin - A Mass Spectrometry Standard,” Mass Spectrom. Rev., vol. 30, pp. 298–320, 2011. doi: 10.1002/mas.20279
[60] K. V Barylyuk, K. Chingin, R. M. Balabin, and R. Zenobi, “Fragmentation of Benzylpyridinium ‘Thermometer’ Ions and Its Effect on the Accuracy of Internal Energy Calibration,” J. Am. Soc. Mass Spectrom., vol. 21, pp. 172–177, 2010. doi: 10.1016/j.jasms.2009.09.023
[61] V. Gabelica and E. De Pauw, “Internal energy and fragmentation of ions produced in electrospray sources,” Mass Spectrom. Rev., vol. 24, pp. 566–587, 2005. doi: 10.1002/mas.20027
[62] J. A. Stolee, Y. Chen, and A. Vertes, “High-energy fragmentation in nanophotonic ion production by laser-induced silicon microcolumn arrays,” J. Phys. Chem. C, vol. 114, no. 12, pp. 5574–5581, 2010. doi: 10.1021/jp906834z
[63] V. Gabelica and E. De Pauw, “Internal energy and fragmentation of ions produced in electrospray sources,” Mass Spectrom. Rev., vol. 24, pp. 566–587, 2005. doi: 10.1002/mas.20027
[64] H. Z. Alhmoud, T. M. Guinan, R. Elnathan, H. Kobus, and N. H. Voelcker, “Surface-assisted laser desorption/ionization mass spectrometry using ordered silicon nanopillar arrays,” Analyst, vol. 139, pp. 5999–6009, 2014. doi: 10.1039/C4AN01391C
[65] S. A. Iakab, P. Ràfols, M. Tajes, X. Correig-Blanchar, and M. García-Altares, “Gold Nanoparticle-Assisted Black Silicon Substrates for Mass Spectrometry Imaging Applications Gold Nanoparticle-Assisted Black Silicon Substrates for Mass Spectrometry Imaging Applications,” ACS Nano, vol. 14, no. 6, pp. 6785–6794, 2020. doi: 10.1021/acsnano.0c00201
[66] R. Sakai et al., “Effects of Carbon Nanowalls (CNWs) Substrates on Soft Ionization of Low-Molecular-Weight Organic Compounds in Surface-Assisted Laser Desorption/Ionization Mass Spectrometry (SALDI-MS),” Nanomaterials, vol. 11, p. 262, 2021. doi: 10.3390/nano11020262
[67] Y. Xiao, S. T. Retterer, D. K. Thomas, J.-Y. Tao, and L. He, “Impacts of Surface Morphology on Ion Desorption and Ionization in Desorption Ionization on Porous Silicon (DIOS) Mass Spectrometry,” J. Phys. Chem. C, vol. 113, pp. 3076–3083, 2009. doi: 10.1021/jp808844f
[68] J. Bian and S. V Olesik, “Ion desorption efficiency and internal energy transfer in polymeric electrospun nanofiber-based surface-assisted laser desorption/ionization mass spectrometry,” Anal. Bioanal. Chem., vol. 412, pp. 1–13, 2020. doi: 10.1007/s00216-019-02315-x
[69] B. N. Walker, J. A. Stolee, D. L. Pickel, S. T. Retterer, and A. Vertes, “Tailored Silicon Nanopost Arrays for Resonant Nanophotonic Ion Production,” J. Phys. Chem. C, vol. 114, pp. 4835–4840, 2010. doi: 10.1021/jp9110103
[70] G. Luo, Y. Chen, H. Daniels, R. Dubrow, and A. Vertes, “Internal Energy Transfer in Laser Desorption/Ionization from Silicon Nanowires,” J. Phys. Chem. B, vol. 110, pp. 13381–13386, 2006. doi: 10.1021/jp0609582
[71] Y. Chen, J. Ding, X. He, J. Xu, and Y. Feng, “Synthesis of tellurium nanosheet for use in matrix assisted laser desorption/ionization time-of-flight mass spectrometry of small molecules,” Microchim. Acta, vol. 185, no. 8, pp. 1–9, 2018. doi: 10.1007/s00604-018-2882-7
[72] A. Vertes, “Soft Laser Desorption Ionization - MALDI, DIOS and Nanostructures,” in Laser Ablation and its Applications, C. R. Phipps, Ed. Santa Fe, New Mexico: Springer, 2007, pp. 505–528.
[73] H. Kawasaki et al., “Platinum Nanoflowers on Scratched Silicon by Galvanic Displacement for an Effective SALDI Substrate,” Chem. A Eur. J., vol. 16, no. 35, pp. 10832–10843, 2010. doi: 10.1002/chem.201001038
[74] S. Dagan, Y. Hua, D. J. Boday, A. Somogyi, R. J. Wysocki, and V. H. Wysocki, “Internal energy deposition with silicon nanoparticle-assisted laser desorption/ionization (SPALDI) mass spectrometry,” Int. J. Mass Spectrom., vol. 283, pp. 200–205, 2009. doi: 10.1016/j.ijms.2009.03.013
[75] M. O. Amin and E. Al-Hetlani, “Development of efficient SALDI substrate based on Au – TiO2 nanohybrids for environmental and forensic detection of dyes and NSAIDs,” Talanta, vol. 233, p. 122530, 2021. doi: 10.1016/j.talanta.2021.122530
[76] X. Jiang, X. Chen, T. Wang, Y. Li, A. Pan, and J. Wu, “Perfluorinated polymer modified vertical silicon nanowires as ultra low noise laser desorption ionization substrate for salivary metabolites profiling,” Talanta, vol. 225, p. 122022, 2021. doi: 10.1016/j.talanta.2020.122022
[77] E. Al-Hetlani, M. O. Amin, M. Madkour, and B. D’Cruz, “Forensic determination of pesticides in human serum using metal ferrites nanoparticles and SALDI-MS,” Talanta, vol. 221, p. 121556, 2021. doi: 10.1016/j.talanta.2020.121556
[78] K. Ng et al., “Ion-Desorption Efficiency and Internal-Energy Transfer in Surface- Assisted Laser Desorption/Ionization: More Implication(s) for the Thermal-Driven and Phase-Transition-Driven Desorption Process,” J. Phys. Chem. C, vol. 119, pp. 23708–23720, 2015. doi: 10.1021/acs.jpcc.5b05957
[79] J. A. Stolee and A. Vertes, “Polarization dependent fragmentation of ions produced by laser desorption from nanopost arrays,” Phys. Chem. Chem. Phys., vol. 13, no. 20, pp. 9140–9146, 2011. doi: 10.1039/C0CP02709J
[80] S. K.-M. Lai, Y.-H. Cheng, H. Tang, and K. Ng, “Silver-Gold alloy nanoparticles as tunable substrates for systematic control of ion-desorption efficiency and heat transfer in surface-assisted laser desorption/ionization,” Phys. Chem. Chem. Phys., vol. 19, no. 20795, pp. 22–25, 2017. doi: 10.1039/C7CP04033D
[81] M. O. Amin and E. Al-Hetlani, “Tailoring the surface chemistry of SiO2-based monoliths to enhance the selectivity of SALDI-MS analysis of small molecules,” Talanta, vol. 200, pp. 458–467, 2019. doi: 10.1016/j.talanta.2019.03.078
[82] N. Li, S. Dou, L. Feng, X. Wang, and N. Lu, “Enriching analyte molecules on tips of superhydrophobic gold nanocones for trace detection with SALDI-MS,” Talanta, vol. 205, p. 120085, 2019. doi: 10.1016/j.talanta.2019.06.085
[83] G. Piret, Y. Coffinier, O. Melnyk, and R. Boukherroub, “Matrix-Free Laser Desorption/Ionization Mass Spectrometry on Silicon Nanowire Arrays Prepared by Chemical Etching of Crystalline Silicon,” Langmuir, vol. 26, no. 8, pp. 1354–1361, 2010. doi: 10.1021/la902266x
[84] W. H. Müller, A. Verdin, E. De Pauw, C. Malherbe, and G. Eppe, “Surface-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging: A Review,” Mass Spectrom. Rev., 2020. doi: 10.1002/mas.21670
[85] Z. Liu, P. Zhang, T. Kister, T. Kraus, and D. A. Volmer, “Ultrathin Homogenous AuNP Monolayers as Tunable Functional Substrates for Surface-Assisted Laser Desorption/Ionization of Small Biomolecules,” J. Am. Soc. Mass Spectrom., vol. 31, pp. 47–57, 2019. doi: 10.1021/jasms.9b00038
[86] R. A. Kruse, X. Li, P. W. Bohn, and J. V Sweedler, “Experimental Factors Controlling Analyte Ion Generation in Laser Desorption/Ionization Mass Spectrometry on Porous Silicon,” Anal. Chem., vol. 73, no. 15, pp. 3639–3645, 2001. doi: 10.1021/ac010317x
[87] L. Qiao and B. Liu, “Nanomaterial-assisted laser desorption ionization for mass spectrometry-based biomedical analysis,” Nanomedicine, vol. 5, no. 10, pp. 1641–1652, 2010. doi: 10.2217/nnm.10.127
[88] T. M. Guinan, P. Kirkbride, P. E. Pigou, M. Ronci, H. Kobus, and N. H. Voelcker, “Surface-Assisted Laser Desorption Ionization Mass Spectrometry Techniques for Application in Forensics,” Mass Spectrom. Rev., vol. 34, pp. 627–640, 2015. doi: 10.1002/mas.21431
[89] A. Y. Lim, J. Ma, Y. Chiang, and F. Boey, “Development of Nanomaterials for SALDI-MS Analysis in Forensics,” Adv. Mater., vol. 24, pp. 4211–4216, 2012. doi: 10.1002/adma.201200027
[90] L. A. McDonnell and R. M. A. Heeren, “Imaging Mass Spectrometry,” Mass Spectrom. Rev., vol. 26, pp. 606–643, 2007. doi: 10.1002/mas.20124
[91] E. R. Amstalden van Hove, D. F. Smith, and R. M. A. Heeren, “A concise review of mass spectrometry imaging,” J. Chromatogr. A, vol. 1217, no. 25, pp. 3946–3954, 2010. doi: 10.1016/j.chroma.2010.01.033
[92] A. R. Buchberger, K. Delaney, J. Johnson, and L. Li, “Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights,” Anal. Chem., vol. 90, pp. 240–265, 2018. doi: 10.1021/acs.analchem.7b04733
[93] T. P. Siegel, G. Hamm, J. Bunch, J. Cappell, J. S. Fletcher, and K. Schwamborn, “Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues,” Mol. Imaging Biol., vol. 20, pp. 888–901, 2018. doi: 10.1007/s11307-018-1267-y
[94] A. Skriba and V. Havlicek, “Mass spectrometry imaging of illicit drugs in latent fingerprints by matrix-free and matrix-assisted desorption/ionization techniques,” Eur. J. Mass Spectrom., vol. 24, no. 1, pp. 124–128, 2018. doi: 10.1177/1469066717728007
[95] T. M. Guinan, C. Della Vedova, H. Kobus, and N. H. Voelcker, “Mass spectrometry imaging of fingerprint sweat on nanostructured silicon,” Chem. Commun., vol. 51, p. 6088, 2015. doi: 10.1039/C4CC08762C
[96] S. J. B. Dunham, J. F. Ellis, B. Li, and J. V. Sweedler, “Mass Spectrometry Imaging of Complex Microbial Communities,” Acc. Chem. Res., vol. 50, no. 1, pp. 96–104, 2017. doi: 10.1021/acs.accounts.6b00503
[97] B. K. Kaletas et al., “Sample preparation issues for tissue imaging by imaging MS,” Proteomics, vol. 9, pp. 2622–2633, 2009. doi: 10.1002/pmic.200800364
[98] R. J. A. Goodwin, “Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences,” J. Proteomics, vol. 75, pp. 4893–4911, 2012. doi: 10.1016/j.jprot.2012.04.012
[99] P. Chaurand, “Imaging mass spectrometry of thin tissue sections: A decade of collective efforts,” J. Proteomics, vol. 75, no. 16, pp. 4883–4892, 2012. doi: 10.1016/j.jprot.2012.04.005
[100] A. Römpp and B. Spengler, “Mass spectrometry imaging with high resolution in mass and space,” Histochem Cell Biol, vol. 139, pp. 759–783, 2013. doi: 10.1007/s00418-013-1097-6
[101] F. Fournelle, E. Yang, M. Dufresne, and P. Chaurand, “Minimizing Visceral Fat Delocalization on Tissue Sections with Porous Aluminum Oxide Slides for Imaging Mass Spectrometry,” Anal. Chem., vol. 92, pp. 5158–5167, 2020. doi: 10.1021/acs.analchem.9b05665
[102] P. Rafols et al., “Assessing the potential of sputtered gold nanolayers in mass spectrometry imaging for metabolomics applications,” PLoS One, vol. 13, no. 12, p. e0208908, 2018. doi: 10.1371/journal.pone.0208908